Exploring the Role of CRISPR/CAS9 Technology and its Utility for Vegetable Improvement
Received Date: March 22, 2025 Accepted Date: April 22, 2025 Published Date: April 25, 2025
doi:10.17303/jacs.2025.4.104
Citation: Pooja Rattan, Neha Kumari, Sandeep Chopra, Anil Bhushan, Dr. Sanjeev Kumar, et al. (2025) Exploring the Role of CRISPR/CAS9 Technology and its Utility for Vegetable Improvement. J Adv Agron Crop Sci 4: 1-13
Abstract
Conventional breeding, which fueled the first Green Revolution, is no longer sufficient to meet the growing challenges of global food security. With the world population projected to surpass 9.7 billion by 2050 and climate change accelerating, we need transformative solutions. Precision breeding and agricultural biotechnology are now pivotal in ushering in a second Green Revolution. Among these technologies, CRISPR/Cas (Clustered Regularly Interspaced Short Palindromic Repeats and associated proteins) has emerged as a game-changing tool in genetic engineering due to its simplicity, precision, scalability, and cost-effectiveness. Enhanced CRISPR-based systems—such as base editing and prime editing—offer even greater versatility, allowing for single-nucleotide modifications without double-strand breaks. These advancements are expanding the frontiers of crop improvement. Genome editing, when combined with next-generation sequencing (NGS) and functional genomics, enables the rapid development of designer crops that are resilient to biotic (pests, pathogens) and abiotic (drought, salinity, temperature) stressors. This is particularly critical for vegetable crops, which are rich in essential micronutrients and contribute significantly to nutritional security. Unlike traditional breeding methods such as selection and backcrossing—which are labor-intensive and time-consuming—CRISPR/Cas9 offers a targeted, efficient alternative to enhance crop yield, quality, disease resistance, stress tolerance, and nutritional value. It facilitates access to untapped germplasm and accelerates the development of elite cultivars by enabling precise gene edits in key regulatory pathways.
Keywords: CRISPR/Cas; Genome Editing; Genetic Engineering; Biotechnology; Mutagenesis
Introduction
One of the most difficult problems that humanity is currently confronting is food security. By 2050, the world's population is predicted to reach 10 billion, meaning that food production must rise by 60–100% worldwide [1]. This issue becomes difficult if we also consider other elements that affect farmers and producers around the world, such as climate change, increased biotic and abiotic stresses, dwindling arable land supply, economic disparities, and differences in political and regulatory systems between countries [2]. Agriculture needs to adopt technological advancements to enhance both the quantity and quality of food, especially as many factors affecting production are difficult or impossible to manage. As a key source of nutrition, vegetable crops play a crucial role in food security. Globally, provide essential metabolites, fiber, vitamins, and minerals vital for human nutrition. However, their physiology makes them particularly susceptible to climate change and adverse weather conditions. Consequently, it is crucial to develop new cultivars capable of rapidly adapting to these evolving environmental challenges.
Traditional breeding has long been essential in addressing the growing the need for increased yields. Yet, this method depends relying on current genetic diversity. Moreover, intensive selection processes have narrowed the genetic pool in certain vegetable species, reducing the presence of alleles for further enhancement [3]. Traditional breeding process is highly labor-intensive and labor-intensive, often requiring taking as long as 20 years to develop a novel variety and bring it to commercial cultivation —a significant challenge when there is an urgent need for superior cultivars. While haploidy induction has been known since the 1950s, the adoption of double haploid plants in breeding programs during the 1990s marked a pivotal advancement, substantially accelerating the development of novel varieties [4]. However, this method still depends on the existing genetic pool and has limited applicability to major vegetable crops due to the scarcity of effective double haploid production techniques.
Advancements and Limitations of Genetic Modification and Genome Editing Technologies in Vegetable Crop Improvement
To overcome these limitations, genetic modification offers a promising alternative. Over the past three decades, different techniques, such as direct DNA uptake, To introduce foreign genetic material into vegetable cells, methods such as particle bombardment and Agrobacterium-mediated transformation have been employed. In 2018, Jaganathan et al. These techniques have been crucial in identifying the roles of genes and creating new vegetable crop features. However, their adoption is constrained by challenges such as inconsistent and occasionally unstable transgene insertions, resistance in certain key vegetable species, and concerns concerning the incorporation of undesired genetic material from vectors and marker genes. These issues have led to stringent global regulations [5]. Targeted genetic changes have been a potent alternative for improving vegetable crops for more than ten years thanks to site-specific nuclease-based approaches. Because of its ease of use, adaptability, and higher efficiency than previous technologies like Zinc Finger Nucleases (ZFNs) and Transcription Activator-Like Effector Nucleases (TALENs), the Clustered Regularly Interspaced Short Palindromic Repeat Associated Protein System (CRISPR/Cas9) is the tool of choice among these [6].Bottom of Form
Modern agriculture utilizes several breeding methods, such as transgenic breeding, mutation breeding, and genetic engineering (GE)-mediated breeding, to enhance crops. In contrast, traditional techniques like genetic recombination and crossbreeding can take years to introduce beneficial alleles and enhance genetic diversity. Breeding with transgenics, a widely recognized method, accelerates the breeding process by incorporating specific genes into economically valuable elite cultivars via exogenous gene transformation. However, this method creates transgenic variations by inserting foreign DNA into random places within the plant genome. Conversely, GE-mediated breeding is more rapid, accurate, and effective than by directly editing genes or regulatory sequences or by changing DNA and RNA bases within elite kinds, genetic engineering (GE) provides targeted improvements and drastically cuts down on the time required to enhance desired qualities. To more typical techniques such as conventional transgenic breeding, mutation breeding, and crossbreeding. CRISPR/Cas9, also known as clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated protein 9 (Cas9), zinc-finger nucleases (ZFNs), transcription activator-like effector nucleases (TALENs), and mega nucleases (MegN) are examples of current GE technologies [7].
The various techniques involved in Genetic engineering are as follows:
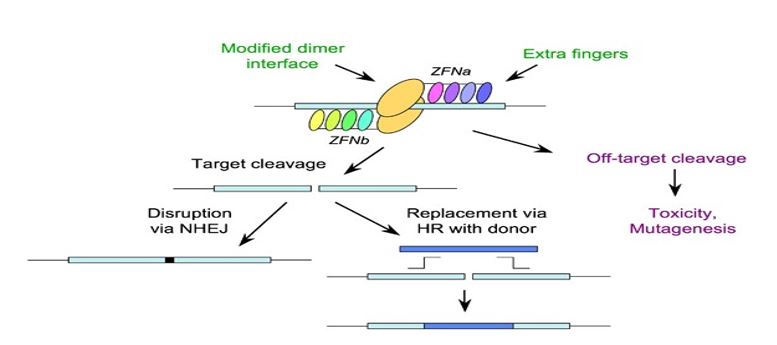
Zinc Finger Nucleases
ZFNs, are made up of two domains: nuclease, which cleaves DNA, and zinc finger proteins, which bind DNA. The nuclease causes a double-strand break at the specified location, while the zinc fingers are made to identify particular DNA sequences. The cell fixes the damage, possibly adding genetic modifications. Moderately precise, but the design of zinc fingers can be complex. The protein's ability to bind to specific sequences depends on the correct alignment of fingers. Designing custom zinc fingers for each target is time-consuming and requires expertise. Each zinc finger has to be synthesized and assembled. ZFNs are effective but have lower efficiency compared to newer methods like CRISPR. There’s also a higher risk of off-target effects. Well-established technique with success in research; used in a few clinical trials.
(TALENs) or Transcription Activator-Like Effector Nucleases
TALENs work similarly to ZFNs but with a different DNA-binding domain. They consist of transcription activator-like effectors (TALEs) that specifically recognize DNA sequences through a repeat structure and a nuclease domain that induces double-strand breaks. TALENs are more precise than ZFNs because the DNA-binding specificity can be altered by changing the repeat sequence, making them versatile in targeting different genes. Designing TALENs is easier than ZFNs because the repeats are more predictable, but the process is still time-consuming. TALENs are relatively efficient, with higher precision than ZFNs, though still not as efficient as CRISPR. Easier design than ZFNs, higher specificity, fewer off-target effects, and more flexible.
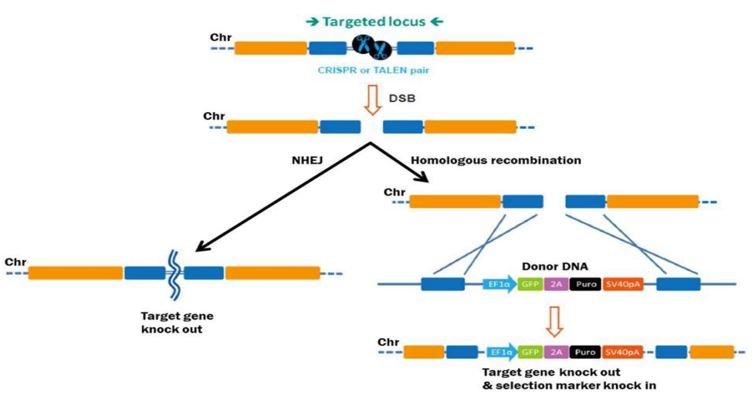
CRISPR/Cas9
A guide RNA (gRNA) is used by CRISPR/Cas9 to lead the Cas9 endonuclease to a particular DNA sequence. At the target site, the Cas9 enzyme subsequently causes a double-strand break. Genetic changes result from the cell's own repair system mending this break. Even though CRISPR/Cas9 can be incredibly accurate, off-target effects can still happen, particularly when guide RNA designs are flawed. Off-target problems have been greatly decreased by recent advancements (such as CRISPR/Cas12 and CRISPR/Cas9 variations). Much easier to design than ZFNs and TALENs because all that needs to be done is create the gRNA that attaches to the target sequence. It may be applied to a wide range of species much more quickly and easily. With little expense and time, CRISPR has greatly improved genome editing and is renowned for its high efficiency. Highly efficient, simple to design, low-cost, and applicable to a wide range of organisms. It has rapidly revolutionized genetic engineering across industries.
CRISPR-CAS9 in Transforming Vegetable Crop Improvement
Using CRISPR-Cas9, the Argonaute 7 gene was deleted to create the first tomato needle-leaf mutant in 2014. Since then, a great deal of study has been done on its possible uses, which include boosting resistance to biotic and abiotic stressors, prolonging shelf life, optimizing plant architecture, and improving fruit quality. The method is currently being researched for a number of crops, including mustard, cabbage, tomatoes, and melons.
Mutating the endogenous phytoene desaturase (PDS) gene and examining the ensuing albino plants is a popular technique to assess gene-editing effectiveness. A recognizable albino phenotype is produced when the PDS gene is disrupted, which lowers the production of carotenoid and chlorophyll. Because Agrobacterium-mediated transformation is easily accessible and gene-editing products produced using this method have no commercial value, tomatoes in particular have emerged as a model crop for exploring CRISPR-Cas9 applications. Crop breeding methods have traditionally involved crossing a superior variety with a donor variety to produce offspring with desirable traits. However, crossbreeding is a lengthy process, often taking 8–10 years to enhance specific traits, such as disease tolerance or resistance, within a species. The selected offspring must undergo multiple backcrosses with the elite variety to eliminate unwanted traits before introducing new desirable traits from the donor variety into the elite line. Mutant breeding, on the other hand, relies on inducing genetic variations through chemical treatments or physical irradiation, typically improving traits over 6–7 years. Transgenic breeding, a simpler and well-established approach, involves introducing external genes into elite varieties, significantly accelerating the process and achieving desired crop improvements in 4–6 years. Genome editing represents a breakthrough in breeding methods, offering precise modification of target genes or regulatory sequences by altering DNA or RNA bases in elite varieties. This technique enables rapid enhancement of specific traits, often within just 2–3 years.
Tomato
Tomato the vegetable crop on which the most CRISPR/Cas research has been conducted. This reflects both the abundance of genomic data and the economic significance of this species. It is also a prime candidate for gene editing due to the availability of extremely effective transformation techniques. Actually, [8] published the first research utilizing the CRISPR/Cas9 technology for any vegetable crop. A. rhizogenes was used to transform the roots of a stable transgenic strain that expressed mGFP controlled by the SCARECROW (SCR) promoter from Solanum lycopersicum.
By altering the mGFP gene's coding region, the initial construct sought to test the system. Different degrees of gfp expression were found in the roots, and the edition events were later verified by restriction enzyme and PCR analysis. By creating a A 19-bp sgRNA similar to eGFP but with only 4 nucleotides different from the mGFP sequence, their second vector evaluated the degree of specificity. The excellent specificity of the sgRNA was confirmed by the absence of any roots exhibiting decreased levels of GFP expression. In order to ascertain whether the function of the gene SHORT-ROOT (SlSHR) was conserved across species, they finally focused on it. Their findings validated the system's ability to carry out gene knockout for functional characterization since they displayed a phenotype that was comparable to that of Arabidopsis. The gene SLARGONAUTE (SLAGO7) was selected because its disruption would lead to a distinct and unique phenotype. According to the data, T0 plants had a significant amount of needle-like leaves. These outcomes demonstrated the efficacy of this gene edition system once more. Research has been conducted on a wide range of subjects since the initial studies validated the idea of utilizing CRISPR/Cas 9 to modify the tomato genome changes made to a number of genes involved in developmental pathways; Targeting the gene RIN in 2015, Ito et al. produced plants with incomplete fruit ripening. CLV3 and SPG5, which are involved in meristematic proliferation and flowering repression, respectively, were altered to produce lines with larger fruit because of aberrant meristem growth and plants that bloomed early because they were less sensitive to long days [9,10].
Mutations in the SlAGL6, which caused plants to produce fruit without seeds. As reported in the second publication, the deletion of SlIAA9 led to plants with modified leaf shapes and the expected seedless fruit. This also resulted in the successful generation of mutant lines with higher levels of aminobutyric acid. [11], lycopene content [12], and a longer shelf life [13] have been reported in relation to significant agronomical features. Research on tomatoes has also focused on several biotic and abiotic stressors. [14] found that when the gene SlMAPK3 was knocked out, the resulting lines had reduced resistance to drought stress. Breeding could benefit from these findings identifying the conserved function by choosing people with high expression of this gene. In order to address the serious issue of plant infections and the vulnerability of certain elite cultivars to them, the gene SlMlo1 was knocked out in order to give tomatoes resistance to powdery mildew. The results showed that this approach was successful in generating pathogen-resistant lines [15]. In the same way. The Moneymaker tomato variety, which is resistant to the bacterial speck disease caused by Pseudomonas syringae pv. tomato. They produced shortened JAZ2 variants devoid of the C-terminal Jas domain by concentrating on altering the SlJAZ2 gene. This modified variety's resistance to necrotrophs was unaffected, although it did exhibit decreased bacterial penetration through the stomata. Along with focusing on desired phenotypic and agronomic characteristics, scientists have also searched for novel uses for this technology. In place of traditional mutagenesis techniques like EMS or fast neutron, [16] suggested using pooled CRISPR/Cas9 libraries to create mutant populations. Their findings demonstrated both high efficiency in producing such mutations and good specificity on the targeted genes.
Cucumber
Cucumber was the first member of the Cucurbitaceae family to successfully use the CRISPR-Cas9 system for gene editing [17]. The researchers targeted two specific sites of the elF4E gene to enhance broad viral resistance. While they achieved virus resistance in cucumbers, the process had low efficiency. Subsequently, [18] aimed to generate gynoecious cucumber lines through CRISPR-Cas9-mediated mutagenesis of the CsWIP1 gene. They used the more resilient CsU6 promoter to increase the system's efficiency. In contrast to the wild-type plants, the resulting T0 mutants had a gynoecious phenotype, which included smaller leaves and exclusively female flowers on the top nodes.
Watermelon
Tian et al. successfully altered this species' CIPDS genes in 2016 to produce knockout mutations. Although plantlets with the anticipated albino phenotype were produced by the disturbance, these lines eventually died as a result of poor shoot development. In a subsequent study, [19] targeted the Acetolactate Synthase (ALS) gene in an attempt to produce watermelon plants that are resistant to herbicides. To confer herbicide resistance, they employed a point mutation technique, specifically creating a C to T alteration in one of the gene's codons. Forty-five of the 199 plants that were created had the necessary mutation. In contrast to the wild-type plants, which were badly impacted by the herbicide Trigonuron, the homozygous mutant lines exhibited total resistance, showing no damage after 14 days.
Potato
In Europe and certain regions of the Americas, it is currently regarded as a staple crop and it has grown to be a highly significant crop globally. The current breeding initiatives place a larger premium on the requirement for cultivars that can improve their nutritional value and adapt to climatic change. This crop has a great chance to benefit from the application of gene edition technology. [24] described the CRISPR/Cas9 technology in potatoes for the first time. To alter the StALS1 gene, they employed diploid and tetraploid types. Three out of the four main events showed more than two types of mutations at a single ALS gene, suggesting that somatic mutations were most prominent in the diploid context. In contrast, four of the five individuals with a tetraploid background exhibited only one type of mutation. The transmission rates of single-targeted mutations ranged from 87% to 100%, and these mutations were inherited through the germlines of both backgrounds. [26] utilized CRISPR-Cas9 to create knockouts in the potato's StIAA2 gene, which is crucial for shoot morphogenesis. Their study demonstrated that this technique was successful in inducing directed mutations for gene functional characterization, as they obtained homozygous and heterozygous plants with different mutations. In another study, [25] targeted the ACETOLACTATE SYNTHASE I (ALSI) gene in potatoes to reduce their susceptibility to ALS-inhibiting herbicides. Their approach involved incorporating a repair template into their vector to facilitate the use of homologous recombination for obtaining modified events.
Additionally, they employed a Geminivirus Replicon (GVR) system and Agrobacterium as transformation techniques. It's interesting to note that their findings demonstrated that only the plants produced via the GVR approach had a greater resistance to herbicides. [28] knocked down the GBSS gene to improve the nutritional content and starch quality. In up to 2% of the regenerated lines, mutations were achieved in all four alleles in a single transfection. Building on these results, [29] utilized CRISPR-Cas9 ribonucleoproteins (RNPs) to deliver the target gene to potato protoplasts, employing a DNA-free genome editing approach. All modified lines were free of transgenes, and RNA-induced mutations were detected in up to 9% of the cases. Unintentional insertions were found at the cut site in over 80% of the shoots with confirmed modifications; however, these insertions were within the expected range of DNA delivery.
Carrot
Carrots are one of the most economically important vegetable crops and a vital species in biotechnology. They are vulnerable to genetic transformation mediated by Agrobacterium and have been crucial in the development of plant tissue culture techniques. However, because of their biennial reproductive cycle, out-crossing tendencies, and the significant consequences of inbreeding depression, carrot research has progressed more slowly than that of other crops. Nevertheless, [20] used the CRISPR-Cas9 system to show targeted mutagenesis in carrots. The researchers used multiplex CRISPR-Cas9 vectors that expressed two single-guide RNAs (gRNAs) to target the flavanone-3-hydroxylase (F3H) gene in order to decrease the production of anthocyanins. They also evaluated the editing effectiveness of three codon-optimized SpCas9 variants (AteCas9, zCas9, and Cas9p) in order to improve the technique. With an astounding 90% mutation rate, the results demonstrated that AteCas9 was the most effective. The F3H gene's crucial function in the biosynthetic pathway that produces anthocyanins was verified by the darkening of calli caused by disruption of the gene.
Lettuce
It is highly important crop with a short life cycle, contributing to a multibillion-dollar industry worldwide. Due to the availability of extensive genomic data, it is considered an ideal candidate for genome editing research. [22] conducted the first study exploring the use of the CRISPR-Cas system in this species. [23] further advanced this research by successfully creating targeted genome modifications in several plant species, including lettuce. Ribonucleoproteins (RNPs) were delivered straight into protoplast cells to accomplish this. The goal was to cause mutations in the BIN2 gene, which functions as a negative regulator in the signaling pathway for brassinosteroids. Analysis showed that the mutant allele might be passed on to the progeny when complete plants were successfully regenerated. The LsNCED4 gene, which controls the thermo-inhibition of lettuce seed germination, was another focus of the team. When this gene was disrupted, the seed's ability to withstand higher germination temperatures significantly improved. More than 70% of the seeds in a trial including 368 T2 plants and 47 primary transformants (T1) were able to germinate, indicating the potential of this genetic change to improve seed germination under stressful circumstances 37°C.
Chicory
One significant vegetable crop that is well-known for generating secondary metabolites with possible medical uses is chicory. Additionally, the food sector uses it, especially in the manufacturing of inulin, a sugar replacement. Chicory is widely grown, thus there is increasing interest in genetically altering it to increase its nutritional value and productivity. [21] investigated the application of targeted mutagenesis based on CRISPR/Cas9 in chicory to accomplish particular genetic alterations. In order to induce a distinct albino phenotype, they concentrated on the CiPDS gene. Depending on the technique, which included direct delivery into protoplasts and Agrobacterium-mediated transformation, they were able to successfully recover albino plants by targeting this gene at rates varying from 4.5% to 31.3%. These results highlight the potential of CRISPR/Cas9 in chicory research, suggesting that such techniques could be further developed to enhance the nutraceutical value of the plant in the future.
Crispr Technology Help in Developing Crops that are more Resilient to Climate Change
CRISPR technology can help develop crops that are more resilient to climate change by enabling precise, rapid, and targeted genetic modifications that enhance tolerance to a range of environmental stresses. By editing genes associated with drought, heat, salinity, and flooding tolerance, CRISPR allows crops such as rice, maize, and wheat to better withstand extreme weather conditions and water scarcity, which are becoming more frequent due to global warming for example, CRISPR has been used to knock out specific genes in rice to improve salt and drought tolerance without compromising yield or growth. The technology can also bolster resistance to diseases and pests whose prevalence and distribution are shifting with climate change, reducing crop losses and the need for chemical pesticides Additionally, CRISPR can be used to improve nutrient use efficiency, allowing crops to thrive with less fertilizer, which not only supports productivity under stress but also reduces agriculture’s environmental footprint Other applications include extending shelf life and reducing food waste, which is critical as climate change increases post-harvest losses. Overall, CRISPR’s speed, accuracy, and versatility make it a powerful tool for developing climate-smart crops that can help secure food supplies in a changing world.
Regulatory Aspects and Public Acceptance of Crispr-Edited Crops
The regulatory landscape for CRISPR-edited crops is complex and evolving, with significant variation across regions. In the United States, the USDA’s APHIS requires developers to notify authorities before advancing gene-edited crops, reflecting a cautious approach, while the SECURE rule exempts certain CRISPR modifications that could be achieved through conventional breeding from stringent regulation, and the FDA and EPA oversee food safety and environmental impacts based on product characteristics rather than editing methods. In the European Union, CRISPR-edited crops are still largely regulated under strict GMO frameworks, making commercialization difficult, though recent policy shifts aim to ease regulations for some gene-editing techniques by enhancing transparency and patent disclosure requirements. Other countries, including China, Thailand, Uruguay, New Zealand, and India, have updated or clarified regulations to support genome-edited crops, often exempting those without foreign DNA from GMO regulations and biosafety assessments, with China notably approving several CRISPR-edited crops for nationwide cultivation36. Intellectual property disputes further complicate commercial access and licensing, influencing innovation and adoption globally. Public acceptance of CRISPR-edited crops varies widely, shaped by cultural, ethical, and informational factors; while some regions are increasingly receptive, skepticism persists, especially where GMOs have faced resistance, underscoring the need for transparent communication about safety and benefits, as well as robust, science-based regulatory frameworks and public education to foster trust and sustainable adoption.
Discussion
As the global demand for food increases, the agribusiness sector must rely on innovative technologies to address the challenges associated with genome editing. Traditional breeding methods are slow and can take years to bring superior cultivars to market, while the public's resistance to transgenic options limits their widespread use [47]. As a result, sequence-specific nucleases such zinc finger nucleases (ZFNs) and transcription activator-like effector nucleases (TALENs) have become viable substitutes for crop improvement. However, there were restrictions on their application and design. Because of its accessibility, simplicity, and accuracy, CRISPR/Cas9-mediated mutagenesis has rapidly emerged as a considerably superior tool in crop improvement [48,49]. This strategy can be used for a number of things, including gene knockdowns, point mutation introduction, mutagenized library creation, and homologous recombination template insertion [50].
The CRISPR/Cas9 system has certain disadvantages despite its efficacy, including the possibility of off-target mutations because of its poor selectivity. Furthermore, not all Cas9 proteins function as well in various crops, which limits its utility [51]. Furthermore, when developing CRISPR-based technologies, government laws and legislation pertaining to gene-edited plants must be considered. For instance, public opinion and research levels in that region have been greatly impacted by the European Union's position on CRISPR [52]. How the technology is incorporated into plant cells will also determine how well CRISPR works in agriculture. The majority of recent studies have employed transformation methods based on Agrobacterium, which could result in two important consequences for genetically modified organisms (GMOs). First, selection genes may be used during the vector generation process, and second, vector DNA may permanently integrate into the host genome. Off-target mutations may increase if the sgRNA is poorly designed or shares a lot of similarities with other genomic areas. Thus, effective and transient delivery methods are essential for enhancing CRISPR's functionality.
Researchers are investigating techniques such as magnetoception, lipofection, and nonoperation for transfection into single cells as alternatives to conventional approaches. About 80% of study studies have concentrated on tomatoes and potatoes, despite the fact that CRISPR technology has not yet been extensively used to the genome editing of vegetable crops. Due in significant part to their significance as staple crops in many regions of the world, tomatoes and potatoes are the focus of genome editing research. Because they can be subjected to genetic transformation, molecular delivery, and regeneration procedures, these species are perfect for genetic research [1]. The advancement of genome editing has been hampered by the greater resistance of other significant vegetable crops, such pepper and onion, to genetic alteration. Developing efficient techniques for molecular delivery and regeneration is necessary to make genome editing a feasible option for these and related crops.
Fortunately, it has been shown that ribonucleoproteins can effectively induce mutations when they are delivered directly into cells [28]. This strategy might be a useful means of avoiding the requirement for in vitro regeneration. Without a question, CRISPR/Cas9-mediated genome editing has the potential to soon transform commercial and research approaches to improving vegetable crops. We think that a new era in food production may be approaching with the correct mix of laws, implementation plans, intellectual property protection, and sustained investment. CRISPR/Cas has immense potential in plant genomics, especially in addressing both biotic (pest and disease-related) and abiotic (environmental stress such as drought, salinity, and temperature extremes) stresses. As genome editing technology advances, CRISPR/Cas is expanding its application to various areas. It can now precisely modify plant genes by editing individual nucleotides, altering amino acids, and even "knocking out," "knocking in," or "knocking up" specific genes. By using CRISPR/Cas to target key genes that regulate a plant's response to these stresses, it is possible to enhance crop growth, yield, and resilience. This approach broadens the genetic resources available, ultimately helping to develop crops that can better withstand environmental challenges and meet the growing global demand for food.
Conclusion
CRISPR/Cas9 is revolutionizing vegetable crop improvement by enabling targeted, efficient, and rapid development of varieties with improved yield, quality, stress resistance, and nutritional value. Continued advances in genome sequencing, regeneration techniques, and regulatory clarity will further expand its impact in the vegetable sector.
- Jaganathan D, Ramasamy K, Sellamuthu G (2018) CRISPR for crop improvement: an update review. Frontiers in Plant Science, 9: 985
- Ma X, Zhu Q, Chen Y (2016) CRISPR/Cas9 platforms for genome editing in plants: developments and applications. Molecular Plant Biology, 9: 961-74.
- Khalil AM (2020) The genome editing revolution: Review. Journal of Genetic Engineering and Biotechnology, 18: 68.
- Kelliher T, Starr D, Richbourg L (2017) MATRILINEAL, a sperm-specific phospholipase, triggers maize haploid induction. Nature, 542: 105–9.
- Waltz E (2018) With a free pass, CRISPR-edited plants reach market in record time. Nature Biotechnology, 36: 6–7.
- Zhang H, Zhang J, Lang Z (2017) Genome editing principles and applications for functional genomics research and crop improvement. Critical Review Plant Science, 36: 291–309.
- Zhang D, Hussain A, Manghwar H, Xie K, Xie S, Zhao S, Larkin RM, Qing P, Jin S, Ding F (2020) Genome editing with the CRISPR-Cas system: An art, ethics and global regulatory perspective. Plant Biotechnology Journal, 18: 1651–69.
- Ron M, Kajala K, Pauluzzi G (2014) Hairy root transformation using Agrobacterium rhizogenes as a tool for exploring cell type-specific gene expression and function using tomato as a model. Plant Physiology, 166: 455–69.
- Xu C, Liberatore KL, MacAlister CA (2015) A cascade of arabinosyltransferases controls shoot meristem size in tomato. Nature Genetics, 47: 784–92.
- Soyk S, Müller NA, Park SJ (2017) Variation in the flowering gene SELF PRUNING 5G promotes day-neutrality and early yield in tomato. Nature Genetics, 49: 162–8.
- Li R, Li R, Li X (2018a) Multiplexed CRISPR/Cas9-mediated metabolic engineering of γ-aminobutyric acid levels in Solanum lycopersicum. Plant Biotechnology Journal, 16: 415–27.
- Li X, Wang Y, Chen S (2018c) Lycopene is enriched in tomato fruit by CRISPR/Cas9-mediated multiplex genome editing. Frontiers in Plant Science, 9: 1–2.
- Yu Q, Wang B, Li N (2017) CRISPR/Cas9-induced targeted mutagenesis and gene replacement to generate long-shelf life tomato lines. Scientific Reports UK, 7: 11874.
- Wang L, Chen L, Li R (2017) Reduced drought tolerance by CRISPR/Cas9-mediated SlMAPK3 mutagenesis in tomato plants. Journal of Agricultural and Food Chemistry, 65: 8674–82.
- Nekrasov V, Wang C, Win J (2017) Rapid generation of a transgene-free powdery mildew resistant tomato by genome deletion. Scientific Reports UK, 7: 1–6.
- Jacobs TB, Zhang N, Patel D (2017) Generation of a collection of mutant tomato lines using pooled CRISPR libraries. Plant Physiology, 174: 2023–37.
- Chandrasekaran J, Brumin M, Wolf D et al (2016) Development of broad virus resistance in nontransgenic cucumber using CRISPR/Cas9 technology. Molecular Plant Pathology, 17: 1140–53.
- Hu B, Li D, Liu X (2017) Engineering non-transgenic gynoecious cucumber using an improved transformation protocol and optimized CRISPR/Cas9 system. Molecular Plant, 10: 1575–8.
- Tian S, Jiang L, Cui X (2018) Engineering herbicide-resistant watermelon variety through CRISPR/Cas9-mediated base-editing. Plant Cell Reports, 37: 1353–6.
- Klimek M, Oleszkiewicz T, Lowder LG (2018) Efficient CRISPR/Cas9-based genome editing in carrot cells. Plant Cell Reports, 37: 575–86.
- Bernard G, Gagneul D, Alves Dos Santos (2019) Efficient genome editing using CRISPR/Cas9 technology in chicory. International Journal of Molecular Sciences, 20: 1155.
- Woo JW, Kim J, Kwon SI (2015) DNA-free genome editing in plants with preassembled CRISPR-Cas9 ribonucleoproteins. Nature Biotechnology, 33: 1162–4.
- Bertier LD, Ron M, Huo H (2018) High-resolution analysis of the efficiency, heritability, and editing outcomes of CRISPR/Cas9-induced modifications of NCED4 in lettuce (Lactuca sativa). G3 Genes, 8: 1513.
- Butler NM, Atkins PA, Voytas DF (2015) Generation and inheritance of targeted mutations in potato (Solanum tuberosum L.) using the CRISPR/Cas system. PLoS ONE, 10: e0144591.
- Butler NM, Baltes NJ, Voytas DF (2016) Geminivirus-mediated genome editing in potato (Solanum tuberosum L.) using sequence-specific nucleases. Frontiers in Plant Science, 7: 1045.
- Wang S, Zhang S, Wang W (2015) Efficient targeted mutagenesis in potato by the CRISPR/Cas9 system. Plant Cell Reports, 34: 1473–6.
- Zhou X, Zha M, Huang J (2017) StMYB44 negatively regulates phosphate transport by suppressing expression of PHOSPHATE1 in potato. Journal of Experimental Botany, 68: 1265–81.
- Andersson M, Turesson H, Nicolia A (2017) Efficient targeted multiallelic mutagenesis in tetraploid potato (Solanum tuberosum) by transient CRISPR-Cas9 expression in protoplasts. Molecular Plant, 36: 117–28.
- Andersson M, Turesson H, Olsson N (2018) Genome editing in potato via CRISPR-Cas9 ribonucleoprotein delivery. Physiologia Plantarum, 164: 378–84.
- Brooks C, Nekrasov V, Lippman ZB (2014) Efficient gene editing in tomato in the first generation using the clustered regularly interspaced short palindromic repeats/CRISPR-Associated9 system. Plant Physiology, 166: 1292–7.
- Ito Y, Nishizawa A, Endo M (2015) CRISPR/Cas9-mediated mutagenesis of the RIN locus that regulates tomato fruit ripening. Biochemical Journal, 467: 76–82.
- Cermák T, Baltes NJ, Cegan R (2015) High-frequency, precise modification of the tomato genome. Genome Biology, 16: 232.
- Xu RF, Li H, Qin RY, Wang L, Li L, Wei PC, Yang JB (2014) Gene targeting using the Agrobacterium tumefaciens-mediated CRISPR-Cas system in rice. Molecular Plant, 11: 3928.
- Pan C, Ye L, Qin L (2016) CRISPR/Cas9-mediated efficient and heritable targeted mutagenesis in tomato plants in the first and later generations. Scientific Reports, 6: 24765.
- Hayut SF, Bessudo CM, Levy AA (2017) Targeted recombination between homologous chromosomes for precise breeding in tomato. Nature Communications, 8: 15605.
- Klap C, Yeshayahou E, Bolger AM (2017) Tomato facultative parthenocarpy results from SIAGAMOUS-LIKE 6 loss of function. Plant Biotechnology Journal, 15: 634–47.
- Shimatani Z, Kashojiya S, Takayama M (2017) Targeted base editing in rice and tomato using a CRISPR-Cas9 cytidine deaminase fusion. Nature Biotechnology, 35: 441–3.
- Nonaka S, Arai C, Takayama M (2017) Efficient increase of G-aminobutyric acid (GABA) content in tomato fruits by targeted mutagenesis. Scientific Reports, 7: 7057.
- Ueta R, Abe C, Watanabe T (2017) Rapid breeding of parthenocarpic tomato plants using CRISPR/Cas9. Scientific Reports, 7: 507.
- Li R, Zhang L, Wang L (2018b) Reduction of tomato-plant chilling tolerance by CRISPR−Cas9-mediated SlCBF1 mutagenesis. Journal of Agricultural and Food Chemistry, 66: 9042–51.
- Dahan T, Filler S, Melamed C (2018) Efficient in planta gene targeting in tomato using geminiviral replicons and the CRISPR/Cas9 system. Plant Journal, 95: 5–16.
- Deng L, Wang H, Sun C (2018) Efficient generation of pink-fruited tomatoes using CRISPR/Cas9 system. Journal of Genetics and Genomics, 45: 51–4.
- Ortigosa A, Gimenez S, Leonhardt N (2019) Design of a bacterial speck resistant tomato by CRISPR/Cas9-mediated editing of SlJAZ2. Plant Biotechnology Journal, 17: 665–73.
- Tomlinson L, Yang Y, Emenecker R (2019) Using CRISPR/Cas9 genome editing in tomato to create a gibberellin-responsive dominant dwarf DELLA allele. Plant Biotechnology Journal, 17: 132–40.
- Tian S, Jiang L, Gao Q (2016) Efficient CRISPR/Cas9-based gene knockout in watermelon. Plant Cell Reports, 36: 399–406.
- Tian S, Jiang L, Gao Q, Zhang J, Zong M, et al. (2017) Efficient CRISPR/Cas9-based gene knockout in watermelon. Plant Cell Reports, 36: 399–406.
- Gupta RM, Musunuru K (2014) Expanding the genetic editing tool kit: ZFNs, TALENs, and CRISPR-Cas9. Journal of Clinical Investigation and studies, 124: 4154–61
- Xie K, Yang Y (2013) RNA-guided genome editing in plants using a CRISPR–Cas system. Molecular Plant, 6: 1975–83
- Arora L, Narula A (2017) Gene editing and crop improvement using CRISPR-Cas9 system. Frontier Plant Science, 8: 1932
- Bortesi L, Fischer R (2015) The CRISPR/Cas9 system for plant genome editing and beyond. Biotechnology Advances, 33: 41–52.
- Belhaj K, Chaparro A, Kamoun S (2015) Editing plant genomes with CRISPR/Cas9. Current Opinion in Biotechnology, 32: 76–84
- CURIA Court of Justice of the European Union (2018) https://curia.europa.eu/jcms/upload/docs/ application/pdf/2018-07/cp180111en.pdf. Accessed 7 Jan 2019

Tables at a glance
Figures at a glance