Evaluation of the mtDNA Cytochrome b gene by PCR-RFLP in three Iranian Horse Breeds
Received Date: November 27, 2023 Accepted Date: December 27, 2023 Published Date: December 30, 2023
doi: 10.17303/jbb.2023.1.106
Citation: Akbar Oghalaie, Milad Oghalaie, Mahmoud Eshagh Hosseini, Mohammad Mehdi Eshagh Hosseini, Fatemeh Kazemi-Lomedasht (2023) Evaluation of the mtDNA Cytochrome b gene by PCR-RFLP in three Iranian Horse Breeds. J Biotechnol Biol 1: 1-7
Abstract
Cytochrome b is one of the proteins of complex III of oxidative phosphorylation that only codes by mitochondrial DNA (mtDNA). The Cytochrome b gene provides suitable phylogenic information in different levels of taxonomy. In this research, 219 blood samples of Kurdish, Iranian Arab, and Turkemin horse breeds were collected from four provinces in North West of Iran. A 590-bp segment comprising the conserved region of the cytochrome b gene of mtDNA was amplified and screened by five restriction enzymes (TaqI, RsaI, BamHI, EcoRI, and HaeIII). Digestion with BamHI enzyme showed two alleles, A (590 bp fragment) and B (106 and 484 bp fragments). The A allele was the most frequent (0.976) among the three breeds. On the other hand, the other restriction endonucleases revealed no polymorphism. Results of the current study strengthened the hypothesis that the Iranian native horse is the ancestor of all horses. Our results for the first time in Iran showed that there were not any differences among three Iranian horse breeds with respect to this part of the mitochondrial genome.
Keywords: Iranian Horse; Mitochondrial DNA; Cytochrome b gene; PCR-RFLP
Introduction
Historical evidence suggests that Iranian native horses are the ancestor of all horses. The most prominent indigenous horse breeds in Iran are Turkemin, Arab and Kurd. The Iranian Arab breed is gradually improving its position mainly due to compilation of a Studbook, close supervision, and holding horse-racing matches. The Turkemin breed is well-preserved for racing purposes under a good management system [1]. On the other hand, the Kurd horse breed exceeds other breeds of horses regarding the most important characteristics in long distance running and ability to live in mountainous areas and walk in rocky conditions. The Kurdish horse is also known by different names, because of differences in their physical appearance, color and location, such as Jaff, Afshari, Sanjabi, and Kalhor [2]. The horse is greatly admired and well-positioned in the religion and culture of Iranian people, which provided further motivation for horse-breeding and racing. Native horses are able of adapting to their environment and withstanding disease and other pressures, while the exotic horses are trained for racing. The total numbers of the existing Iranian Arab, Turkemin, and Kurd breeds in Iran were estimated to be approximately 1000-1200, 300-400, and 1000-1500, respectively, in 2004. Today, the number of purebred horses has decreased because of uncontrolled breeding with unknown breeds [1]. Nevertheless, due to the diversity of fauna, Iran is an important center of purebred horses [1,3]. Mitochondrial DNA (mtDNA) analysis has often been used in evolutionary studies [4,5]. Sequence polymorphisms of mtDNA have been analyzed to elucidate the phylogenetic relationships among and within several animal species [5]. Research conducted on mitochondrial DNA variation in horses utilized PCR-RFLP analysis to investigate a 590-base pair segment of the cytochrome b gene across six distinct horse breeds. The findings unveiled a significant level of diversity within this genomic segment, facilitating the categorization of the studied horses into four distinct types [6]. In the current study, we used PCR-RFLP to compare mtDNA variations observed among Iranian horse breeds by analyzing the 590-bp segment of the cytochrome b gene.
Materials and Methods
Blood Sampling and DNA Extraction
The study was based on a total of 219 registered horses from the west of Iran as follows: 171 Kurdish horses, 21 Iranian Arab horses, 15 Turkemin horses, and 12 horses of unknown breeding. Blood samples were obtained from each horse using vacuum tubes containing EDTA for an anticoagulant, from June 2007 to May 2008. Genomic DNA was isolated using a standard salt extraction method.
PCR-RFLP
The 590-bp fragment of the cytochrome b gene was amplified using sequences of primers previously described [6]. The PCR was performed in a reaction volume of 20 µL containing approximately 50 ng of genomic DNA, 40 pmol of each primer, and 4 µL of Master Mix 5X [7]. The PCR reaction mixture was heated to 96oC for 2 min followed by 35 cycles, each consisting of 1 min at 94oC, 1 min at 55oC, and 1 min at 72oC. The final extension was carried out at 72oC for 10 min. The PCR products were digested with five restriction endonucleases (RsaI, BamHI, HaeIII, EcoRI, and TaqI) under conditions recommended by the manufacturers (Fermentas, Vilnius, Lithuania). The choice of these restriction endonucleases was based on previous studies [6]. Digested fragments were separated using 1.8% agarose or 8% polyacrylamide gel electrophoresis and visualized with UV illumination of ethidium bromide or silver staining, respectively.
Statistical Analyses
The number of individuals of each genotype within each breed was used to determine Hardy-Weinberg equilibrium, genotypic frequencies, and allelic frequencies. Population genetic indices, such as the effective number of alleles (Ne), observed heterozygosity (Ho), expected heterozygosity (He), and inbreeding coefficient (FIS), were estimated using Popgene 32 software version 1.32 (11) and the polymorphic information content (PIC) [8].
Results and Discussion
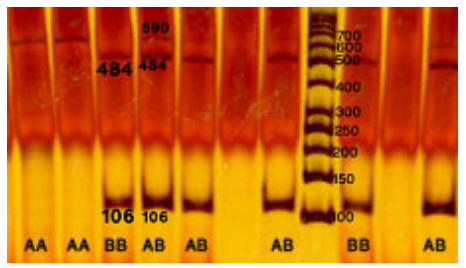
Due to the important roles of the cytochrome b gene in identification of maternity and lineage, this region is considered to be a reliable marker for the history of horse breeding and evolutionary routes. This study found polymorphism patterns in all of the horse breeds investigated. The PCR products digested by RsaI, TaqI, HaeIII, and EcoRI revealed no polymorphism in any of the breeds. Figure 1 shows typical horse mtDNA cleavage patterns of the 590-bp DNA fragment in the cytochrome b gene digested by BamHI restriction endonulease. Digestion of the 590-bp PCR product with the BamHI restriction endonulease resulted in two DNA bands (484 and 106 bp) for homozygote BB and three bands (590, 484, and 106 bp) for the AB heterozygote. The DNA amplified from homozygous AA animals remained undigested with BamHI. When the individual PCR products from the 219 horses were digested with other restriction endonuleases (TaqI, RsaI, EcoRI, and HaeIII), no polymorphism was observed.
No RsaI polymorphisms in a partial fragment of the cytochrome b gene in Thoroughbred, Hokkaido, Kiso, and Misaki breeds of horses [6]. Analysis of the cytochrome b RFLP-RsaI polymorphism showed that among the Jeju, Tsushima, and Thoroughbred breeds of horses, only the Jeju breed exhibited polymorphism [7]. It has been reported that examination of a 1350-bp segment comprising the entire cytochrome b gene by RsaI endonuclease in five Greek horse breeds presented three alleles [9]. The horses were monomorphic for TaqI, whereas HaeIII revealed two alleles.
In our study, PCR products digested by BamHI showed two different morphs in Iranian horse breeds. Morph A was the most frequent pattern, which showed one fragment (590 bp), whereas Morph B was the less frequent pattern, demonstrating the digested fragments (484 bp and 106 bp). Analysis of a 590-bp fragment of the cytochrome b gene by BamHI enzyme in Thoroughbred, Korean, and Mongolian breeds [6] and in Jeju and Thoroughbred horses [7] demonstrated two morphs similar to those seen in our study. Comparison of our results with those in the literature [6,7], which showed no differences between polymorphisms of this region of the genome in Iranian horses and that of Thoroughbred horses, strengthens the hypothesis that Iranian horse breeds are ancestors of all Thoroughbred horses in the world.
Until now, no other papers have been found in the literature related to analysis of the cytochrome b gene in Iranian horse breeds and this is the first report regarding the examination of this region of the horse genome in Iran. Although it may be difficult to distinguish one breed from the others using only our PCR-RFLP results, this PCR-RFLP would be useful for maternity testing in the Iranian horse registry.
The allelic and genotypic frequencies of products digested by BamHI are summarized in Table 1. The frequency of the A-allele was much higher than that of the B-allele in the analyzed populations. The distribution of genotypes and alleles for the Iranian Arab breed followed the HardyWeinberg rule (P > 0.05). The rest of breeds were not in Hardy-Weinberg equilibrium (P < 0.01), indicating that there were some probable components of disequilibrium, such as nonrandomized breeding, selection, migration, and genetic drift.
Table 2 summarizes the values for Ho , He , Ne , PIC and FIS. The Ho and He values in the Iranian Arab breed were identical, but differed slightly in the Kurdish breed. He in the Turkemin breed and in the horses of unknown breeding was considerably higher than Ho . The Ne value for the Turkemin breed was higher than in the other breeds, indicating that both alleles are present in the Turkemin population. The Polymorphism Information Content (PIC) serves as a parameter that reflects the extent of informativeness associated with a genetic marker. Following the criteria of (3), the Turkemin breed revealed medium genetic diversity (0.25 < PIC < 0.50), but the other breeds showed low genetic diversity (PIC < 0.25). The fixation index (FIS) serves as an indicator of either a deficiency or excess of heterozygotes. Negative FIS values signify an excess of heterozygotes (indicative of outbreeding), while positive values denote a deficiency of heterozygotes (suggestive of inbreeding), as compared to the expectations under Hardy–Weinberg equilibrium. The Iranian Arab breed exhibited a negative FIS, whereas the values of this index were highly positive for both the Turkemin horses and for the horses of unknown breeding, implying a deficiency of heterozygosity in the latter two groups.
Conclusion
Our results underscore the utility of mitochondrial DNA (mtDNA) as a valuable instrument for conducting population genetic studies in Iranian horses. The comprehensive analysis of extensive datasets encompassing diverse breeds enhances our comprehension of evolutionary and domestication processes in equines. Additionally, molecular data can be leveraged to gauge and preserve ample genetic diversity within and across breeds, serving as instrumental tools in the formulation and implementation of effective management strategies for breed conservation.
Authorship
Akbar Oghalaie; Methodology, Experiment, Writing - Review & Editing.
Milad Oghalaie; Writing - Review & Editing,
Mahmoud Eshagh Hosseini; Writing - Review & Editing
Mohammad Mehdi Eshagh Hosseini; Writing - Review & Editing.
Fatemeh Kazemi Lomedasht; Review & Editing
Acknowledgment
The authors acknowledge Pasteur Institute of Iran for supporting this article. We gratefully acknowledge the contribution of the Sanandaj organization of Jihad-e-Agriculture, especially the Deputy of Improvement of Animal Products, Mr. Kh. Jafari, for his official support and Mr. A. M. Mahmoodi for collection of the horse blood samples.
- Kohler-Rollefson I (2004) Farm animal genetic resources: Safeguarding national assets for food security and trade.
- Tavanaei-Manesh H, Dalir-Naghadeh B (2010) Electrocardiographic parameters in purebred Kurd horse. Journal of Animal and Veterinary Advances 9: 2698-703
- Takasu M, Hiramatsu N, Tozaki T, Kakoi H, Nakagawa T, Hasegawa T, et al. (2012) Genetic characterization of the endangered Kiso horse using 31 microsatellite DNAs. Journal of Veterinary Medical Science 74: 161-6.
- Cozzi MC, Strillacci MG, Valiati P, Bighignoli B, Cancedda M, Zanotti M (2004) Mitochondrial D-loop sequence variation among Italian horse breeds. Genetics Selection Evolution 36: 1-10.
- Kim G, Kim S, Oh M (1999) PCR-RFLP of the cytochrome b genes in the Cheju native horses. KOREAN JOURNAL OF GENETICS 21: 29-34.
- Ishida N, Hasegawa T, Mukoyama H, Oyunsuren T (1996) PCR‐RFLP analysis of the cytochrome b gene in horse mitochondrial DNA. Animal Genetics 27: 359-63.
- Han SH, Kim JH, Song JH, Oh JH, Oh YS, Jung YH, et al. (2004) Polymorphism of the mtDNA cytochrome B and NADH dehydrogenase 6 genes in Tsushima and Jeju native horses. Genes & Genomics 26: 1-7.
- Botstein D, White RL, Skolnick M, Davis RW (1980) Construction of a genetic linkage map in man using restriction fragment length polymorphisms. American journal of human genetics 32: 314.
- Apostolidis AP, Alifakiotis T, Mamuris Z, Karkavelia E (2000) PCR‐RFLP analysis of mitochondrial DNA cytochrome b gene among Greek horse breeds. Italian Journal of zoology 67: 159-62.

Tables at a glance
Figures at a glance