Cloning and Prokaryotic Expression of 17β-HSD Reproductive Gene in the Ivory Shell, Babylonia areolata
Received Date: November 18, 2024 Accepted Date: December 18, 2024 Published Date: December 21, 2024
doi: 10.17303/jbb.2024.2.103
Citation: Yangting Lin, Chenlin Guo, Meiying Wang, Nan Cai, Guo-Fang Jiang (2024) Cloning and Prokaryotic Expression of 17β-HSD Reproductive Gene in the Ivory Shell, Babylonia areolata. J Biotechnol Biol 2: 1-9
Abstract
17β-HSDs belong to the short-chain dehydrogenase/reductase (SDR) superfamily of retinol, and 17β-HSD11 is the catalytic enzyme in the final step of sex steroid hormone synthesis, converting testosterone into dihydrotestosterone to reduce androgen activity and affect gender differentiation in mollusks. However, the 17β-HSD11 gene is not specifically expressed in gonads. In this study, we screened a transcriptome database from the ivory shell Babylonia areolata and identified one reproductive 17β-HSD11 gene. The full length of this gene was found to be 2,720 bp with a cDNA sequence of 1,296 bp encoding a protein consisting of 432 amino acids with a relative molecular weight of 47 kDa. Bioinformatics analysis revealed that the encoded protein belongs to hydrophobic transmembrane stabilizing proteins. We amplified the protein coding region of the 17β-HSD11 gene using PCR technology and constructed a recombinant protein expression vector for it using pET28a. After screening and identification of bacterial colonies, we successfully introduced the recombinant protein expression vector and extracted the plasmid for induction expression experiments. The induced protein showed similar relative molecular weight as expected, and our experiment determined that an optimal IPTG inducer concentration for inducing protein expression was at 0.3 mM.
Keywords: Babylonia areolata; 17β-HSD11 Gene; Gene Cloning; Prokaryotic Expression
Introduction
Gastropoda is the largest class in the phylum Mollusca, and the processes of sex determination and differentiation are very diverse. In the hermaphroditic population, different scholars have different views on sex determination and differentiation, and the factors affecting sex determination mainly include karyotype and chromosomes, genetic factors, and environmental factors. A few species, such as the sea snail Janthina janthina, can change sex through physical contact with the same species [1]. In mollusks, sexual differentiation is controlled by neuromodulation, and neurohormones act on membrane receptors to initiate a series of intracellular events that regulate gene transcription [2]. The structure and sequence of Hdh-GnRH in Pacific abalone Haliotis diversicolor in mollusks were found to be similar to gonadotropin-releasing hormone (GnRH) analogue control pathways, and the results showed that the structure and sequence of Hdh-GnRH in Pacific abalone were strictly conserved with those of GnRH-like peptides in other invertebrates, and the specific expression of GnRH in ganglia was significantly increased during the mature stage of Pacific abalone, indicating that GnRH was related to mollusc reproduction [2].
17β-HSDs belong to the retinol short-chain dehydrogenase/reductase (SDR) superfamily, and 17β-HSD11 is a catalytic enzyme in the final step of the sex steroid hormone synthesis process, which oxidizes testosterone to androstenedione, which can reduce androgen activity [4]. It has been found that the 17β-HSD11 gene is expressed in all parts of the body, and the 17β-HSD11 gene is highly expressed in the gonads, kidneys, livers and other tissues related to metabolism in humans and mice [5], and the 17β-HSD11 gene is mainly expressed in the gonads in mollusks. Other subtypes of the 17β-HSD family have other functions in addition to the regulation of estrogen and male hormones, and the 17β-HSD10 and 17β-HSD12 genes are also involved in the metabolism of fatty acids in the body [6-7], and the 17β-HSD10 gene is also involved in the metabolism of neurosteroids.
In this paper, the 17β-HSD11 gene was cloned to enrich the genes related to the reproduction of the ivory shell, Babylonia areolataha, which is of great significance for the sex determination of B. areolataha. In addition, the bioinformatics analysis of 17β-HSD11 protein in this paper is helpful to further understand the structural characteristics of 17β-HSD11 protein, and also provides a certain reference value for the subsequent qualitative and semi-quantitative analysis of 17β-HSD11 protein.
Materials and methods
Laboratory Animals
In June 2022, the same snail was divided into 5 tissues (foot, salivary glands, snout, gills, and gonads) and bagged separately, and the males and females were distinguished according to the color of the gonads, and then the 5 tissues of the same snail were put into the same sealed bag, quickly placed in liquid nitrogen, and put them together in the -80°C refrigerator for later use after all the samples were processed.
Extraction of total RNA
RNA was extracted in a sterile bench using the Ultrapure RNA Kit, 3 μL was used for agarose gel electrophoresis to detect whether the RNA isolation was successful, and the remaining RNA was immediately placed in a -80 °C freezer to avoid RNA degradation.
Reverse transcription and primer design of 17β-HSD reproductive genes
The RNA extracted was reverse transcribed into cDNA using a full-form gold reverse transcription kit (Wuhan Jinkairui Bioengineering Co., Ltd., China). The 17β-HSD gene sequence screened from the second-generation transcriptome data of B. areolata was compared with NCBI to obtain the conserved region of the gene, and the primer sequences (DFL1F: GCATTTTCGTTCCGCCCATC, DFL1R: AGACAACAGGAGTTCCCCCACT) were designed for the conserved region by Primer premier 5.0 software, and their annealing temperature was 54°C.
PCR amplification and purification
Taq enzyme 2× Easy Taq PCR SuperMix (Beyotime Biotechnology Co., Ltd., Shanghai, China) was amplified using the cDNA of B. areolate as a template, and the total reaction system was 20 μL (Table 1). The reaction was divided into three parts: pre-denaturation: 94 °C, 3 mins; annealing extension: 35 cycles, 94 °C, 30 s; 54.6 °C 30 s; 72 °C, 83 s; final extension: 72 °C, 10 mins. After amplification, the target band can be detected directly by agarose gel electrophoresis, or the PCR product can be stored at 4 °C for later use. The amplified band was cut under ultraviolet light, the gel with the target band was put into a 1.5 mL centrifuge tube, weighed with an electronic balance, purified with a biogel recovery and purification kit, purified by agarose gel electrophoresis to detect whether the purification was successful, and the purified product was stored at 4 °C before being sent to the company for sequencing.
Construction of 17β-HSD gene prokaryotic expression vector
After the competent cells were thawed in an ice bath, 50 μL was taken, 2.5 μL of pET-28a no-load plasmid was added, gently mixed and placed in an ice bath for 30 min, then heat shocked by water bath for 42 °C for 45 s, and quickly transferred to an ice bath for 2 mins to introduce the no-loaded plasmid into the competent cells, 500 μL of Luria-Bertani liquid medium without kanamycin was added to the centrifuge tube, mixed and placed on a shaker at 37 °C, 180 rmp for 1 h. Centrifuge 5 mL of bacterial solution at 12,000 rmp for 2 mins, and discard the supernatant. The PET-28A plasmid was extracted using a biotech plasmid extraction kit.
BamH I and Xho I were selected as enzyme cleavage sites, and primers for prokaryotic expression in the conserved region of 17β-HSD gene were designed, and PCR amplification and agarose gel electrophoresis were performed. The FlyCut® BamH I and FlyCut® Xho I reagents (TransGen Biotech Co., Ltd.,China) were loaded with full-formula gold according to the enzyme digestion system, and the target gene and plasmid were double-digested, and the digestion system is shown in Table 1 and 2. After loading, mix well and put into a PCR machine at 37 °C for 25 mins. Add 6× DNA Lodding Buffer to dilute to 1 × to inactivate the endonuclease. The inactivated digested product and the undigested target gene and plasmid were verified by agarose gel electrophoresis to verify whether the enzyme digestion was successful, and the product was purified with a biogel recovery and purification kit.
The digested 17β-HSD gene and plasmid were ligated by T4 DNA Ligase reagent (TransGen Biotech Co., Ltd) to construct a plasmid expression vector. The reaction system was constructed, mixed after sample loading, and placed in a thermal cycler at 25 °C for 30 mins.
Transformation of Escherichia coli
After the recombinant plasmid was introduced into the competent cells, 500 μL of LB liquid medium without kanamycin was added to the centrifuge tube, mixed well, and placed on a shaker at 37 °C, 180 rmp for 1 h. On a sterile table, 100 μL of the activated bacterial solution was added to LB solid medium containing kanamycin antibiotic (kanamycin concentration 30 μL/mL), and the bacterial solution was spread with a coating rod, the plate was inverted and sealed with parafilm, and placed in a 37 °C incubator inverted overnight.
Identification of recombinant bacteria
Colony PCR was used to verify the recombinant bacteria and colony PCR reaction system with the colony direct PCR kit (Beyotime Biotechnology Co., Ltd., Shanghai). Firstly, a single colony was picked from the Kanata antibiotic LB solid medium with a sterile pipette, added to 10 ml of liquid medium containing Kanamyces antibiotic LB, and placed in a shaker at 37 °C at 180 rmp, and incubated overnight to expand the strain. Then, the samples were mixed according to the reaction system, and the PCR products were amplified in a thermal cycler, and the PCR products were verified by agarose gel electrophoresis.
In order to further ensure the correctness of the recombinant plasmid, the strains corresponding to the correct PCR bands of the colony were extracted, and the extracted plasmids were sent to the company for Sanger sequencing, and the sequencing results were compared with the expected sequences.
Recombinant protein induction
200 μL of the bacterial solution with correct sequencing results was added to 5 mL of liquid medium containing Kanatella antibiotic LB liquid medium, placed in a shaker at 37 °C, 180 rmp for 3 h, and the control group in this experiment was E. coli containing 200 μL of unloaded plasmid under the same conditions. Then different concentrations of IPTG inducers were added to the bacterial solution: 0 mM, 0.1 mM, 0.2 mM, 0.3 mM, 0.4 mM and numbered, 1 mL of the expanded bacterial solution was taken, centrifuged at 12,000 rpm for 5 mins, and the supernatant was discarded. 50 μL of 2×SDS-PAGE loading buffer was added to the pellet, and the E. coli pellet was completely mixed with the loading buffer by repeated pipetting, and the samples were boiled in boiling water for 10 mins. Centrifugation at 12,000 rpm and 2 minutes were ready for sample loading.
SDS-PAGE assay: A polyacrylamide gel was prepared, a marker was added to the first lane, and protein samples with IPTG concentrations of 0 mM, 0.1 mM, 0.2 mM, 0.3 mM, and 0.4 mM were added to the second to sixth lanes. When the staining solution reaches the bottom 0.5 cm of the gel, stop electrophoresis, remove the gel and rinse it with deionized water for 3 times, and stain it with Coomassie brilliant blue stain solution for 3 h until the gel is completely invisible, pour off the staining solution and add the destaining solution to destain until clear protein bands can be seen. After destaining is complete, observe under a gel imager to determine if the recombinant protein is expressed.
Bioinformatics analysis
The gene sequence information obtained by amplification and splicing comparison in the conserved region was predictively analyzed by relevant bioinformatics analysis software, and the homology of the homologous sequence was analyzed by BLAST software (https://blast.ncbi.nlm.nih.gov/Blast.cgi), and the amino acid homology comparison was carried out.
Results
Gene sequence analysis
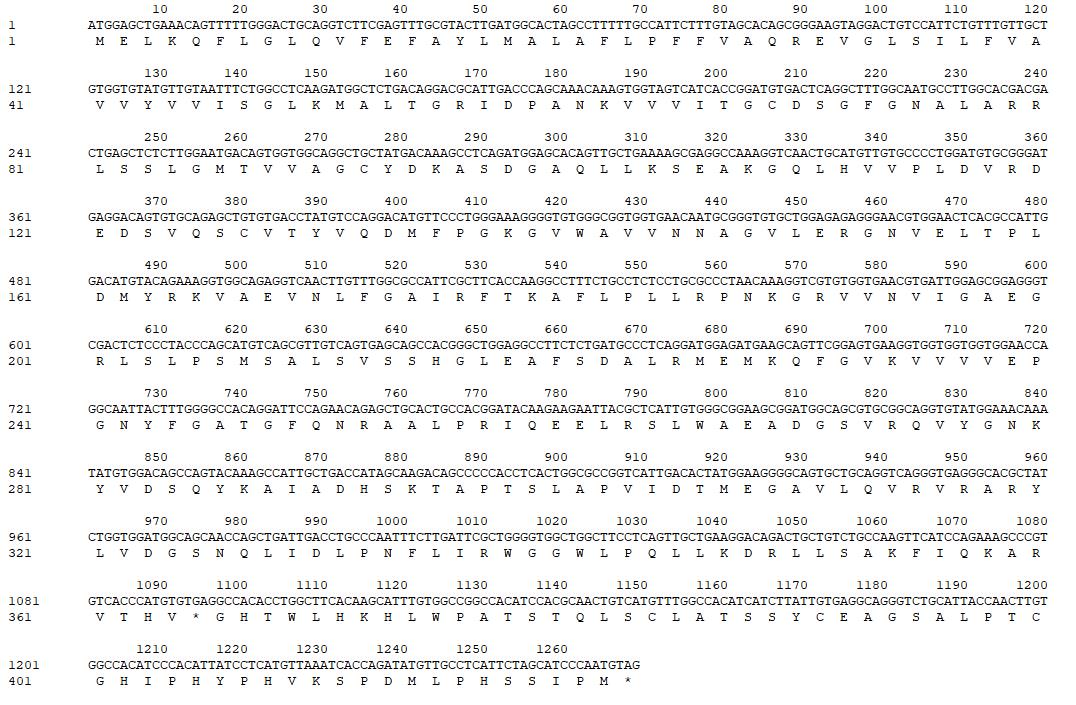
The cDNA sequence of 17β-HSD gene was 1 269 bp in length and encoded 430 aa, as shown in Figure 1.
Analysis of protein properties
The relative molecular weight of 17β-HSD protein was 47 kDa, the number of amino acids was 432, the isoelectric point was 8.95, and the molecular formula was C2119H3351N575O594S19. It contains 20 kinds of amino acids, of which the highest content is leucine Leu (L) (11.6%), followed by valine Val (V) (10.9%), and the lowest content is Trp (W) (1.4%). There were 35 acidic amino acids (Asp+Glu) and 42 basic amino acids (Arg+Lys).
17β-HSD aliphatic index 99.05, total average hydrophilicity 0.146, the protein is hydrophobic protein, the instability index is 36.36, the GRP (guruprasad-reedy-pandit) method is used to determine whether the protein is stable, according to the standard instability coefficient (II) greater than 40 is the unstable protein, less than 40 is all stable proteins except RNaseA, Therefore, the protein is a stable protein.
Amino acid sequence alignment of closely related species
The 8 amino acid sequences of 17β-HSD were downloaded from the closely related species and compared with the amino acid sequences obtained in this study. The results were shown in Figure 2, with high homology among the 9 sequences.
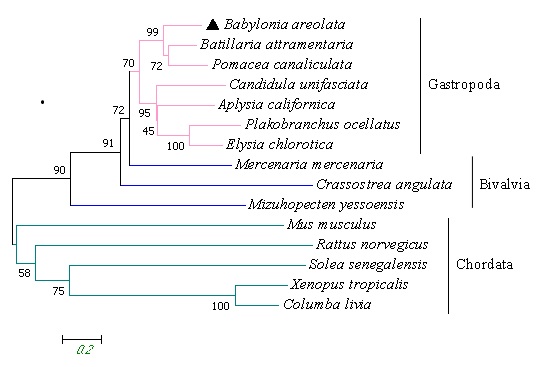
Phylogenetic tree analysis
The phylogenetic tree relationship of 17β-HSD11 protein is shown in Figure 3, and the results show that the amino acid sequence of 17β-HSD11 of B. areolate is clustered with Batillaria attramentaria and Pomacea canaliculata in Gastropoda, indicating that they are closely related. Then, the 17β-HSD11 protein was clustered with other animals in the mollusk, and finally clustered with vertebrates into a large group, and the confidence in clustering was high, indicating that the 17β-HSD11 protein in the animal kingdom had high homology and genetic relationship.
Discussion
The 17β-HSD11 gene of B. areolata is the conserved structural characteristics of SDR superfamily genes. Protein structure prediction revealed that the β helix contains disulfide bonds, and the Cu/Zn sites in each subunit can be responsible for catalyzing the superoxide disproportionation reaction [8]. At present, there are nine kinds of amino acids that can be phosphorylated, among which serine, threonine, and tyrosine are particularly common as phosphorylation sites [9], which is consistent with the prediction of phosphorylation sites of B. areolata 17β-HSD11 protein in this paper, and the phosphorylation of proteins can induce changes in protein conformation, thereby regulating the nature and intensity of protein-protein interactions, and then coordinating different reaction pathways [9]. The results of amino acid sequence alignment showed that the 17β-HSD protein belonged to the 11 isoform and had high homology with the amino acid sequences of other species, indicating that the 17β-HSD11 protein was relatively conserved in the evolutionary process. Through phylogenetic tree analysis, it was found that the genetic relationship between B. areolata and Pomacea canaliculata was closer and had good collinearity. Compared with the snail, the research on the heredity of the snail is relatively backward, and the genetic information of the snail can be used to strengthen the genetic research of the snail, which is of great significance for the discovery of the new gene of the snail.
In order to allow the recombinant protein to be successfully expressed, in addition to the selection of cells in the genus group, it is also necessary to consider the conditions for induced expression, the recombinant protein usually exists in the form of inclusion bodies because it cannot be folded and curled spontaneously in Escherichia coli, and protein folding may also occur in proteins that form soluble proteins at lower induction temperatures [10], but according to the Niu’s research [11], this result is not fixed, and the results are not much different from those at 37 °C and 16 °C, so the induced protein was selected at 37 °C. The results showed that the optimal induction conditions were obtained at 37 °C after adding 0.3 mM IPTG for 5 h, but the solubility of the protein was not analyzed, and the soluble protein could be obtained by reducing the expression temperature in the future, so as to conduct more in-depth research on 17β-HSD11 protein, and also provide a certain basis for the further study of its application.
The 17β-HSD11 gene is active in the gonads and digestive glands of mollusks such as spinnaker mussels, scallops, and abalone [13-15], and the 17β-HSD11 gene has similar biological functions to vertebrate homologous genes in mollusks, that is, it affects sex differentiation by controlling the synthesis of sex steroids. 17β-HSD11 is used as an oxidase in variegated abalone to reduce androgen activity by oxidizing testosterone to androstenedione [14], while 17β-HSD12 in nine-hole abalone acts as a reductase, which can convert estrone into estradiol and increase estrogen activity [15]. Wang et al.'s study on Moneyfish showed that the increase in the expression of 17β-HSD gene is related to a certain signaling pathway, and 17β-HSD acts on the transport or biotransformation of cholesterol and other substances, making it biosynthesized into steroid hormones to promote gonadal development [16]. It can be seen that sterol hormones are of great significance for the synthesis of sex hormones.
In the 17β-HSD family, subtype 2 is involved in the regulation of steroid hormones, although it belongs to oxidase, it has the same efficiency for oxidizing androgens and estrogen, subtype 7 can not only regulate estrogen and androgen in the body, but also convert yeast sterone into yeast sterols to participate in the regulation of cholesterol metabolism in the body, and subtype 10 begins to be a protein isolated from the central nervous system of patients with Alzheimer's disease, which plays an important role in cognition [4], These suggests that different genes may have unique functions in addition to the common role in a family, and the function of 17β-HSD11 gene can be hypothesized and further validated by referring to the unique functions of other subtypes. Thitiphuree et al.'s study of Ctenophores scallop showed that 17β-HSD11 was most expressed in the liver, suggesting that 17β-HSD11 is not only related to reproduction, but may also be involved in other metabolisms [17]. Although 17β-HSD11 has not been proven to be involved in fatty acid metabolism in molluscs, it has been found in vertebrates in recent years that 17β-HSD11 is involved in catalyzing fatty acid metabolism. Liu et al. found that the N-terminal hydrophobic domain of the 17β-HSD11 protein is required for targeted binding to lipid droplets, while other catalytic sites are not involved in the regulation of lipid droplets, based on the secondary structure analysis of 17β-HSD11 and the expression of truncated proteins encoding the full-length protein of 17β-HSD11 [18]. Although the N-terminal sequence in 17β-HSD11 has weak homology with the N-terminal sequence of the PAT family [19], the specific targeting mechanism of different families of lipid droplet-targeted proteins, such as the HSD family and the PAT family, is unknown [17]. In addition, the sequence similarity between 17β-HSD11 and 17β-HSD13 is 65% [18], and 17β-HSD13 has been found to have a similar role in regulating fatty acid metabolism [10]. Both 17β-HSD11 and 17β-HSD13 are associated with Nonalcoholic fatty liver disease (NAFLD), suggesting that NAFLD can be treated by regulating the expression of these two genes.
- Cahill A E, Juman A R, Pellman-Isaacs A, et al. (2015) Physical and chemical interactions with conspecifics mediate sex change in a protandrous gastropod Crepidula fornicata. Biology Bulletin, 229: 276-81.
- Ketata I, Denier X, Hamza-Chaffai A, et al. (2008) Endocrine-related reproductive effects in molluscs. Comparative Biochemistry and Physiology: Toxicology and Pharmacology, 147: 261-70.
- Li W-D, Min H, Lü W-G, et al. (2015) Involvement of antizyme characterized from the small abalone Haliotis diversicolor in gonadal development. PLoS One, 10: e0135251.
- Su W, Xu H M, Kang J H, et al. (2014) Function of 17β-hydroxysteroid dehydrogenase. Advances in physiological science, 45: 27-31.
- Liu Y, Xu S, Zhang C, Zhu X, Hammad M A, Zhang X, Christian M, Zhang H, Liu P (2018) Hydroxysteroid dehydrogenase family proteins on lipid droplets through bacteria, C. elegans, and mammals. Biochim Biophys Acta Mol Cell Biol Lipids, 1863: 881-94.
- Zhang Y, Wang Q, Ji Y, Zhang Q, Wu H, Xie J, Zhao J (2014) Identification and mRNA expression of two 17β-hydroxysteroid dehydrogenase genes in the marine mussel Mytilus galloprovincialis following exposure to endocrine disrupting chemicals. Environ Toxicol Pharmacol, 37: 1243-55.
- Floyd BM, Drew K, Marcotte EM (2021) Systematic identification of protein phosphorylation-mediated interactions. Journal of Proteome Research, 20: 1359-70
- Sun F, Ren M, Zhao L, et al. (2023) Research progress on the structure and function of membrane proteins regulated by phosphorylation. Journal of Hebei University of Technology, 52: 1-12.
- Sorensen HP, Mortensen KK, et al. (2005) Microbial cell factories biomed central review soluble expression of recombinant proteins in the cytoplasm of Escherichia coli. Microbial Cell Factories, 4: 1-8.
- Niu AJ (2022) Prokaryotic expression, purification and preparation of polyclonal antibody against canine FGF12. Changchun: Jilin Agricultural University.
- Shangguan X (2022) Molecular Identification and functional study of Genes related to sex steroid hormone synthesis in Hyriopsis cumingii. Shanghai: Shanghai Ocean University.
- Liu J (2014) Potential role of sex steroid hormones and 17β-hydroxysteroid dehydrogenase 8 in gonad development in Chlamys farreri. Qingdao: Ocean University of China.
- Gao Y (2010) Identification of structure and function of 17β hydroxysteroid dehydrogenase 12 in Abalone. Beijing: Tsinghua University.
- Zhai HN, Zhou J, Cai ZH (2012) Cloning, characterization, and expression analysis of a putative 17 beta-hydroxysteroid dehydrogenase 11 in the abalone, Haliotis diversicolor supertexta. Journal Steroid Biochemistry Molecular Biology, 130: 57-63.
- Wang W (2017) Cloning, prokaryotic expression and functional verification of gonadotropin gene in Monocarp. Shanghai: Shanghai Ocean University.
- Thitiphuree T, Nagasawa K, Osada M (2019) Molecular identification of steroidogenesis-related genes in scallops and their potential roles in gametogenesis. The Journal of Steroid Biochemistry and Molecular Biology, 18622-33.
- Liu Y L, Xu S, Zhang C Y, et al. (2018) Hydroxysteroid dehydrogenase family proteins on lipid droplets through bacteria, C. elegans, and mammals. Biochimica et Biophysica Acta-Molecular and Cell Biology of Lipids, 1863: 881-94.
- Horiguchi Y, Araki M, Motojima K (2008) Identification and characterization of the ER/lipid droplet-targeting sequence in 17beta-hydroxysteroid dehydrogenase type 11. Archives of Biochemistry and Biophysics, 479: 121-30.
- Yang L, Ding YF, Chen Y, et al. (2012) The proteomics of lipid droplets: structure, dynamics, and functions of the organelle conserved from bacteria to humans. Journal of Lipid Research, 53: 1245-53.


Tables at a glance
Figures at a glance