| Cell Line |
Retrovirus |
Clonogenic Efficiency (Relative to C127 Cells) |
Inhibition of Clonogenicity (Relative to LXSN) |
| MCF7 (n=4) |
LXSN |
2.0% |
N/A |
| ErbB4 |
1.6% |
15 ± 2% |
| ErbB4 Q646C |
0.04% |
97 ± 1% |
| ErbB4 Q646C K751M |
3.3% |
None |
| ErbB4 Q646C V673I |
2.5% |
None |
| ErbB4 Q646C LL783/4AA |
2.4% |
None |
| ErbB4 Q646C LL867/8AA |
0.5% |
70 ± 7% |
| ErbB4 Q646C L985A |
2.9% |
None |
| ErbB4 Q646C D990A |
1.2% |
35 ± 13% |
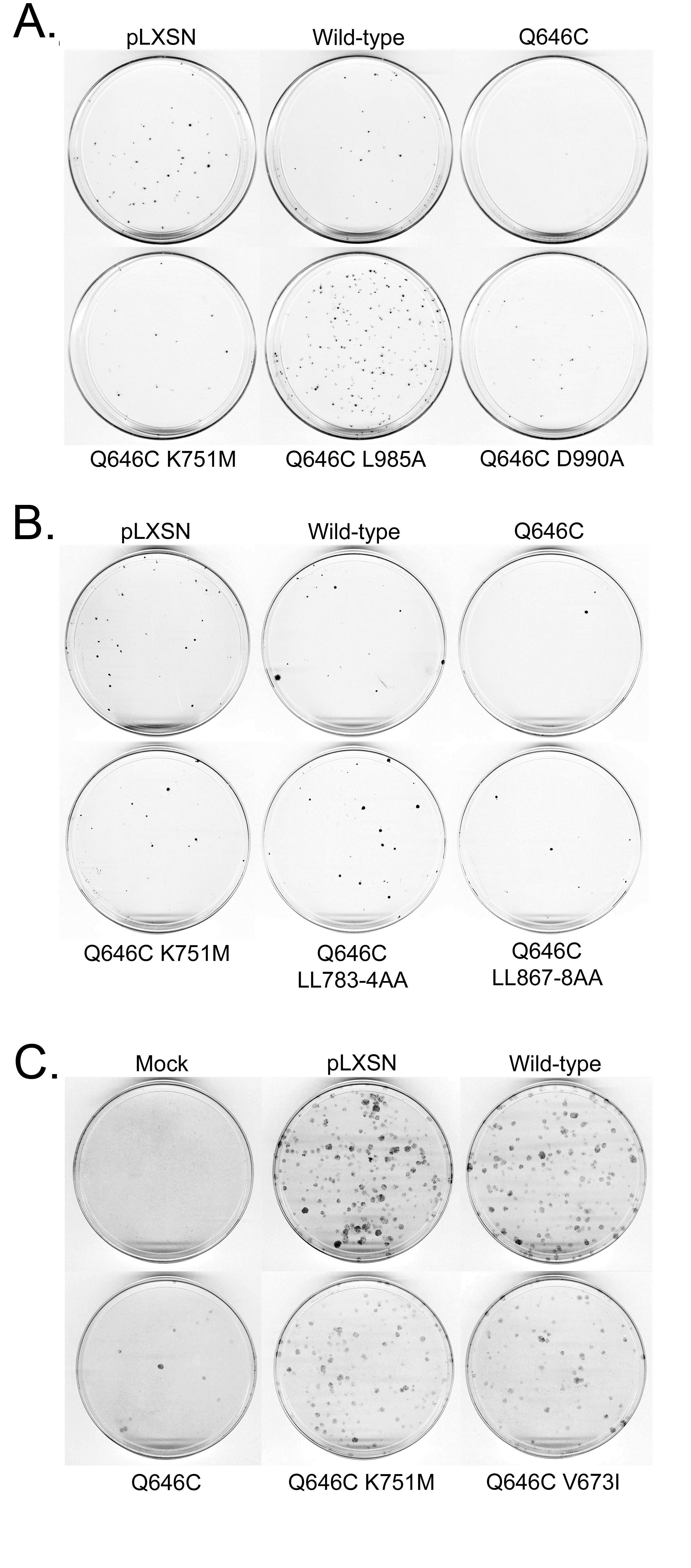
Table 1: The K751M, V673I, LL783/4AA, and L985A mutations markedly disrupt the tumor suppressor activity of the constitutively-active ErbB4 Q646C
mutant in the MCF7 human breast tumor cell line.
| Cell Line |
Retrovirus |
Clonogenic Efficiency (Relative to C127 Cells) |
Inhibition of Clonogenicity (Relative to LXSN) |
| Expt. 1 MCF10A (n=4) |
LXSN |
9.8% |
N/A |
| ErbB4 |
10.0% |
None |
| ErbB4 Q646C |
0.8% |
91 ± 1% |
| ErbB4 Q646C K751M |
8.5% |
None |
| ErbB4 Q646C V673I |
8.5% |
None |
| Expt. 2MCF10A(n=3) |
LXSN |
4.3% |
N/A |
| ErbB4 |
6.2% |
None |
| ErbB4 Q646C |
0.5% |
87 ± 2% |
| ErbB4 Q646C K751M |
9.5% |
None |
| ErbB4 Q646C LL783/4AA |
7.4% |
None |
| ErbB4 Q646C LL867/8AA |
1.8% |
58 ± 6% |
| Expt. 3MCF10A(n=3) |
LXSN |
5.8% |
N/A |
| ErbB4 |
7.1% |
None |
| ErbB4 Q646C |
0.6% |
88 ± 3% |
| ErbB4 Q646C K751M |
7.4% |
None |
| ErbB4 Q646C L985A |
5.8% |
None |
| ErbB4 Q646C D990A |
4.4% |
28 ± 13% |
Table 2: The K751M, V673I, LL783/4AA, and L985A mutations markedly disrupt the tumor suppressor activity of the constitutively-active ErbB4 Q646C mutant in the MCF10A human breast epithelial cell line.
| Cell Line |
Retrovirus |
Clonogenic Efficiency (Relative to C127 Cells) |
Inhibition of Clonogenic Proliferation (Relative to ErbB4 Q646C K751M) |
| DU-145 (n=4) |
ErbB4 Q646C K751M |
7.4% |
N/A |
| ErbB4 Q646C |
0.3% |
95 ± 2% |
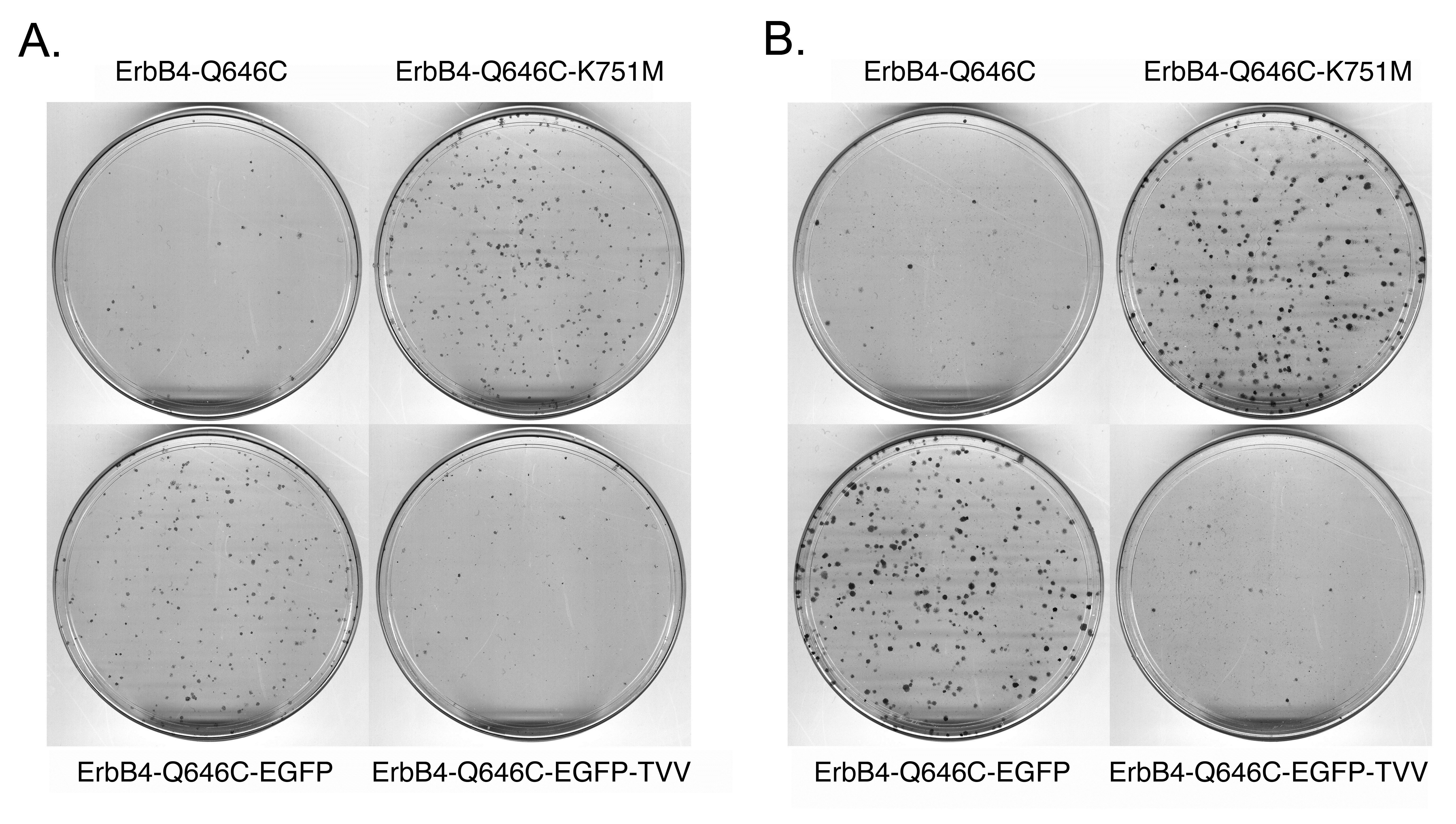
| ErbB4 Q646C EGFP |
6.0% |
15 ± 9% |
| ErbB4 Q646C EGFP-TVV |
1.2% |
83 ± 4% |
| PC-3 (n=3) |
ErbB4 Q646C K751M |
12.0% |
N/A |
| ErbB4 Q646C |
1.4% |
90 ± 3% |
| ErbB4 Q646C EGFP |
10.9% |
15 ± 8% |
| ErbB4 Q646C EGFP-TVV |
1.1% |
89 ± 3% |
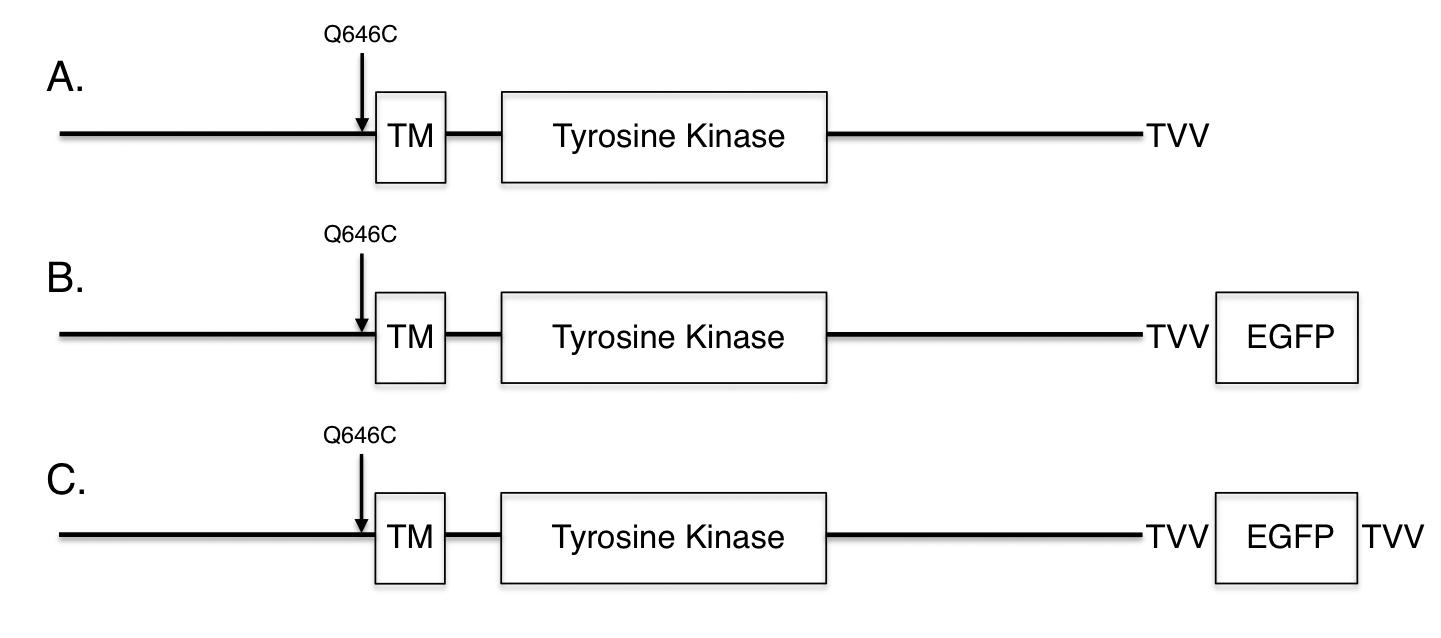
Table 3: Adding a carboxyl-terminal Enhanced Green Fluorescent Protein (EGFP) tag to the constitutively-active ErbB4 Q646C mutant disrupts its tumor suppressor activity, but this deficit is rescued by adding a carboxylterminal Thr-Val-Val (TVV) sequence.
| Cell Line |
Retrovirus |
Clonogenic Efficiency (Relative to C127 Cells) |
Inhibition of Clonogenic Proliferation (Relative to ErbB4 Q646C K751M) |
| DU-145(n=5) |
ErbB4 Q646C K751M |
4.6% |
N/A |
| ErbB4 Q646C EGFP |
5.5% |
None |
| ErbB4 Q646C EGFP-TVV |
0.8% |
82 ± 2% |
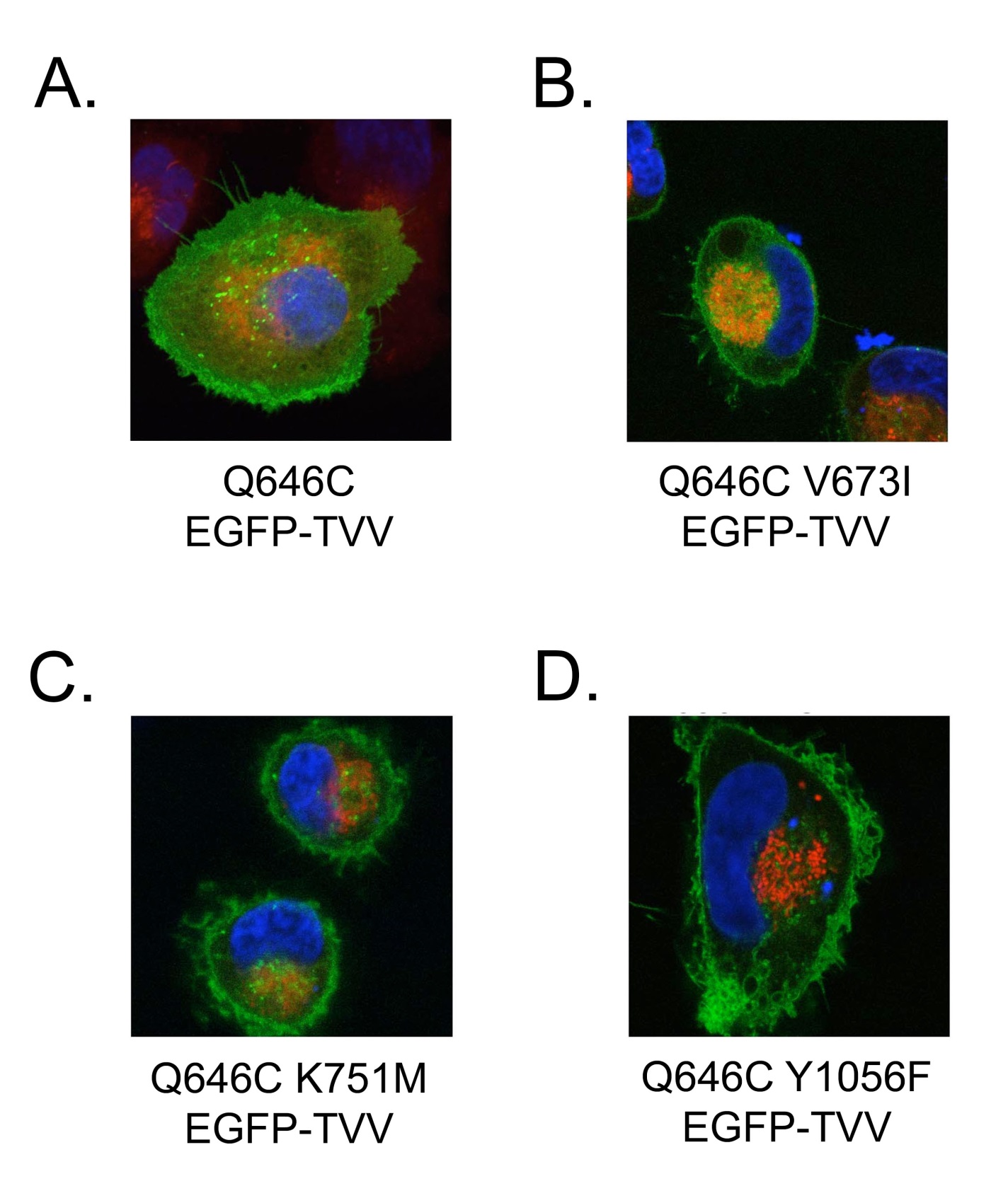
| ErbB4 Q646C K751M EGFP-TVV |
12.8% |
None |
| ErbB4 Q646C V673I EGFP-TVV |
5.2% |
None |
| ErbB4 Q646C LL783/4AA EGFP-TVV |
8.9% |
None |
| ErbB4 Q646C LL867/8AA EGFP-TVV |
0.4% |
92 ± 2% |
| ErbB4 Q646C L985A EGFP-TVV |
2.8% |
35 ± 10% |
| ErbB4 Q646C D990A EGFP-TVV |
1.4% |
69 ± 4% |
| ErbB4 Q646C Y1056F EGFP-TVV |
9.7% |
None |
| PC-3(n=5) |
ErbB4 Q646C K751M |
15.7% |
N/A |
| ErbB4 Q646C EGFP |
24.9% |
None |
| ErbB4 Q646C EGFP-TVV |
2.0% |
89 ± 3% |
| ErbB4 Q646C K751M EGFP-TVV |
22.9% |
None |
| ErbB4 Q646C V673I EGFP-TVV |
8.9% |
28 ± 17% |
| ErbB4 Q646C LL783/4AA EGFP-TVV |
26.1% |
None |
| ErbB4 Q646C LL867/8AA EGFP-TVV |
3.8% |
80 ± 3% |
| ErbB4 Q646C L985A EGFP-TVV |
8.5% |
49 ± 6% |
| ErbB4 Q646C D990A EGFP-TVV |
3.3% |
78 ± 3% |
| ErbB4 Q646C Y1056F EGFP-TVV |
23.4% |
None |
Table 4: The K751M, V673I, LL783/4AA, and Y1056F mutations markedly disrupt the tumor suppressor activity of the constitutively-active ErbB4 Q646C EGFP-TVV construct in the DU-145 and PC-3 human prostate tumor cell lines.