Expression Signature Panel for Brain Tissues and Developmental Stages
Received Date: August 01, 2024 Accepted Date: September 01, 2024 Published Date: September 04, 2024
doi: 10.17303/jcrto.2024.12.305
Citation: Yossi Cohen (2024) Expression Signature Panel for Brain Tissues and Developmental Stages. J Cancer Res Therap Oncol 12: 1-14
Abstract
Gene expression profile (GEP) plays a pivotal role in characterizing various tissues and tumors. However, distinguishing between different brain tissue types and tumors based on gene expression remains a formidable challenge. This difficulty arises due to the existence of over 20 distinct brain tissue types, a similar number of brain tumors, and the rarity of tissue-specific genes.
In this comprehensive meta-analysis, I introduce a novel approach—an expression intensity signature panel specific to brain tissues. Leveraging the characteristic expression patterns of thousands of genes for each brain tissue type and tumor, this reference panel demonstrates remarkable alignment with histological diagnoses. Notably, this alignment persists even when accounting for variations in transcriptomic techniques, platforms, and species.
Furthermore, the application of this reference panel to fetal brain transcriptomes yields intriguing insights. Fetal brain tissues, including the cerebral cortex, cerebellum, hippocampus, and amygdala, maintain an embryonic cellular stage until mid-pregnancy in humans and sheep. Similarly, in mice and rats, this embryonic signature endures into early postnatal life. The transition from a primitive to a mature expression signature occurs rapidly during specific postnatal days in these animals.
Keywords: Brain Tissues; Gene Expression Profile; Tumors; Brain Development; CNS
Introduction
Histological diagnosis remains the cornerstone for determining tumor type and its cell of origin. However, this approach encounters several challenges, including technical complexities, potential loss of tissue-specific markers, and variability in interpretation among different observers. Moreover, classical histopathology alone may not always definitively establish the cell of origin, necessitating alternative approaches such as GEP to subclassify diverse tumors [1-3].
In this meta-analysis, I propose a novel tissue-specific expression intensity signature panel—a valuable reference for correlating with test brain specimens. The foundation of this panel lies in the distinct expression patterns exhibited by thousands of genes associated with each specific brain tissue type. Constructed from 43 different normal and tumor brain transcriptomes, this reference panel selectively incorporates overexpressed probes that characterize each tissue or tumor type. Analogous to identifying an unknown DNA sequence by aligning it with known sequences, this reference panel—comprising representative expression intensity signatures from 43 known histologies—was employed to correlate with the expression of the same probes in test brain specimens. This approach enables us to identify the optimal alignment based on the strongest correlation.
By leveraging this tissue-specific expression intensity signature panel, I aim to enhance diagnostic precision, especially in cases where conventional histopathology alone falls short or to improve the uniformity of specimens for clinical trials.
Methods
To construct the reference tissue-specific expression intensity signature panel, I systematically gathered GEP series from a diverse array of brain tissues spanning 43 histological normal and tumor categories. These GEPs were sourced from the Gene Expression Omnibus (GEO) public repository (GPL570 platform).
For each individual histological brain category (such as the cerebellum, amygdala, and medulloblastoma), I generated representative GEPs by calculating the average expression intensity of each of the 54,615 probes within the selected series. To enhance specificity, I removed probes that did not exhibit overexpression in at least one histological brain category relative to any other. This curation process involved pairwise comparisons of the 43 representative GEPs, resulting in the identification of 4,621 probes that constitute each reference tissue-specific expression intensity signature. These signatures were then correlated with test brain specimens. For additional details, please refer to the Supplementary Methods section.
Subsequently, I validated the reference panel using transcriptomes from independent brain specimens. These transcriptomes are publicly deposited in the GEO database and are also accessible on the BRAINSPAN website: https://www.brainspan.org/static/download.html . The diagnostic precision of the reference panel was compared to that of conventional histological diagnosis.
Availability and Implementation
The alignment, mathematically represented by correlation, between each of the 43 reference tissue-specific expression intensity signature probes and the corresponding 4,621 probes from over 60,000 brain transcriptomes deposited in public repositories is visually presented in the form of an atlas. Researchers can readily access this comprehensive resource for download from the website https://brain-cell - of-origin-atlas.website
The platforms and species analyzed are detailed in Table 1, and the complete list of the 521 datasets analyzed in Supplemental Table 1.
Main
The correlations between the reference tissue-specific expression intensity signature probes and corresponding probes from normal cortical, cerebellar, spinal, thalamic, meningeal, choroid, and pituitary specimens consistently demonstrated their strongest alignments with analogous histological categories.
Specifically
Cerebellar Transcriptomes
Show almost 100% concordance with corresponding cerebellar reference signature probes across all series (refer to Table 1 and the Atlas files).
Cerebral Transcriptomes
Also exhibit remarkably high concordance.
Accumbens and Striatu
Impressively show concordance in specific series (GSE45642(i), GSE87823(i))
Amygdala and Hippocampus
Display variable concordance with corresponding reference expression signatures, with the highest consistency observed in larger series (e.g., GSE86574 and GSE45642 in Atlas I, GSE60862 in Atlas II).
The discrimination between such similar histological tissues is by no means trivial, especially considering the scarcity of known tissue-specific probes (like ZIC2 and GABRA6 in the cerebellum). The limited number of tissue-specific probes is obviously insufficient for fold change discrimination among the 20 closely related brain tissues.
Notably, even with only 8 specimens available per each normal brain category (from GSE86574), which construct nearly the entire reference panel for normal brain tissues, we achieve effective discrimination among different brain categories. This achievement underscores the power and advantage of using expression signatures, each comprised of thousands of probes, over fold change discrimination based on small numbers of highly expressed probes (i.e., >2 folds) for discrimination.
Furthermore, the reference panel effectively differentiates brain tumors from each other and from normal brain specimens, particularly in cases involving immature or embryonic tumors.
While the reference panel effectively discriminates between various brain tissues and tumors, its sensitivity does not extend to distinguishing tumor versus normal tissues of the same cell of origin, such as normal ependyma versus ependymoma. Instead, it primarily reflects the fundamental cell lineage. Indeed, glioma transcriptomes exhibit a less consistent pattern, often overlapping with the reference expression signature of normal cerebral specimens. This observation suggests that the origin of gliomas involves transformed glial cells distinct from neural stem cells or precursor glial cells, as elaborated by some researchers [4-6]. Alternatively, contamination of glioma tumor biopsy specimens by normal cerebral tissue might contribute to this overlap. Ensuring pure tumor samples remains a challenge in clinical practice.
Unfortunately, the available data does not allow to directly examine whether gliomas that best align with normal cerebral reference signature probes exhibit different clinical outcomes.
In the realm of single-cell gene expression profiles (GEPs) for gliomas, diverse cells within the same tumor exhibit alignment with different reference GEPs (as observed in Atlas IV, Atlas V, and Atlas VI). This intriguing finding suggests the potential presence of subclones or the coexistence of cancer stem cells within glioma specimens.
Another noteworthy observation arises from GSE25219 (Atlas II): The expression intensity probes from human brain transcriptomes obtained at 6-8 weeks of gestational age matched the corresponding reference signature probes from embryonic tumors with multilayered rosettes (ETMR). In contrast, those obtained at 9-17 weeks aligned more closely with medulloblastoma or primitive neuroectodermal tumors (PNET). This suggests that the origin of ETMR lies at an earlier embryonic stage than that of medulloblastoma or PNET.
Surprisingly, the reference panel, generated solely from the Affymetrix GPL570 series, demonstrates utility across other microarray platforms and even RNA-seq platforms by matching the corresponding probes. Additionally, the reference expression intensity signatures exhibit remarkable concordance with corresponding test signatures from animal brain transcriptomes, including those from monkeys, mice, rats, and other species (refer to Supplemental Table 1).
Chimpanzee Brain Tissues (Atlas III)
Within the limits of the small number of specimens, chimpanzee cerebellar, cerebral, striatal, thalamic, and pituitary tissues all excellently match the corresponding human reference probes.
The hippocampus exhibits overlapping patterns with the amygdala.
Interestingly, the monkey subiculum part overlaps with cerebral and amygdala reference signatures, while other parts only overlap with the cerebral reference signature.
Mouse Brain Tissues
Cerebellar and Cerebral Tissues, Choroid, and Meninges:
Match the corresponding human reference probes well.
The striatum match varies by series (excellent in GSE13588).
Other Mouse Brain Tissues
Align with the cerebral reference probes, except for the retina, which aligns with the cerebellar probes (GSE33088).
However, in some mouse hippocampal series (GSE62346 and GSE240873), all mouse hippocampal specimens match the reference probes of the amygdala, similar to the pattern observed in monkeys.
This raises doubt about whether the specimens included the same anatomical structures or were perhaps contaminated with cerebral tissue
The same holds true for mouse striatum, which excellently matches the human striatal reference in GSE113842.
Rat Brain Tissues
Show an excellent match for cerebellar, cerebral, spinal, and pituitary probes with their corresponding reference probes.
Rat hippocampal transcriptomes often align with the cerebral reference signature, but in some series (GSE29552), they align with the amygdala reference.
Interestingly, the rat superior olivary complex matches human vestibular nuclei (GSE34003/GSE16764).
Wild Pigs
Hypothalamic and pituitary probes match excellently with the corresponding human reference probes (GSE109155, GSE240433).
The hippocampus and amygdala best match the human cortex reference, but their second-best match is with the hippocampus and amygdala, respectively.
Avian Brain (Singing Bird Taeniopygia guttata)
Area X (part of the avian basal ganglia, shaping the songs of zebra finches and providing insights into human speech disorders) matches both the human striatal and cerebral areas.
HVC, LMA, and RA areas match the cerebral probes only.
In relation to animal brain tumors, the only available series is from mouse transcriptomes. For Mouse Brain Tumors, an excellent match exists between medulloblastoma (GSE24628, GSE11859, GSE65888, GSE69359, GSE112699, GSE36594, GSE29192) and retinoblastoma with their corresponding reference probes (GSE29685).
New Insights into Brain Developmental Stages Using the Reference Panel
In addition to analyzing normal and tumor brain transcriptomes, the reference panel has also been applied to fetal brain series, yielding fascinating results.
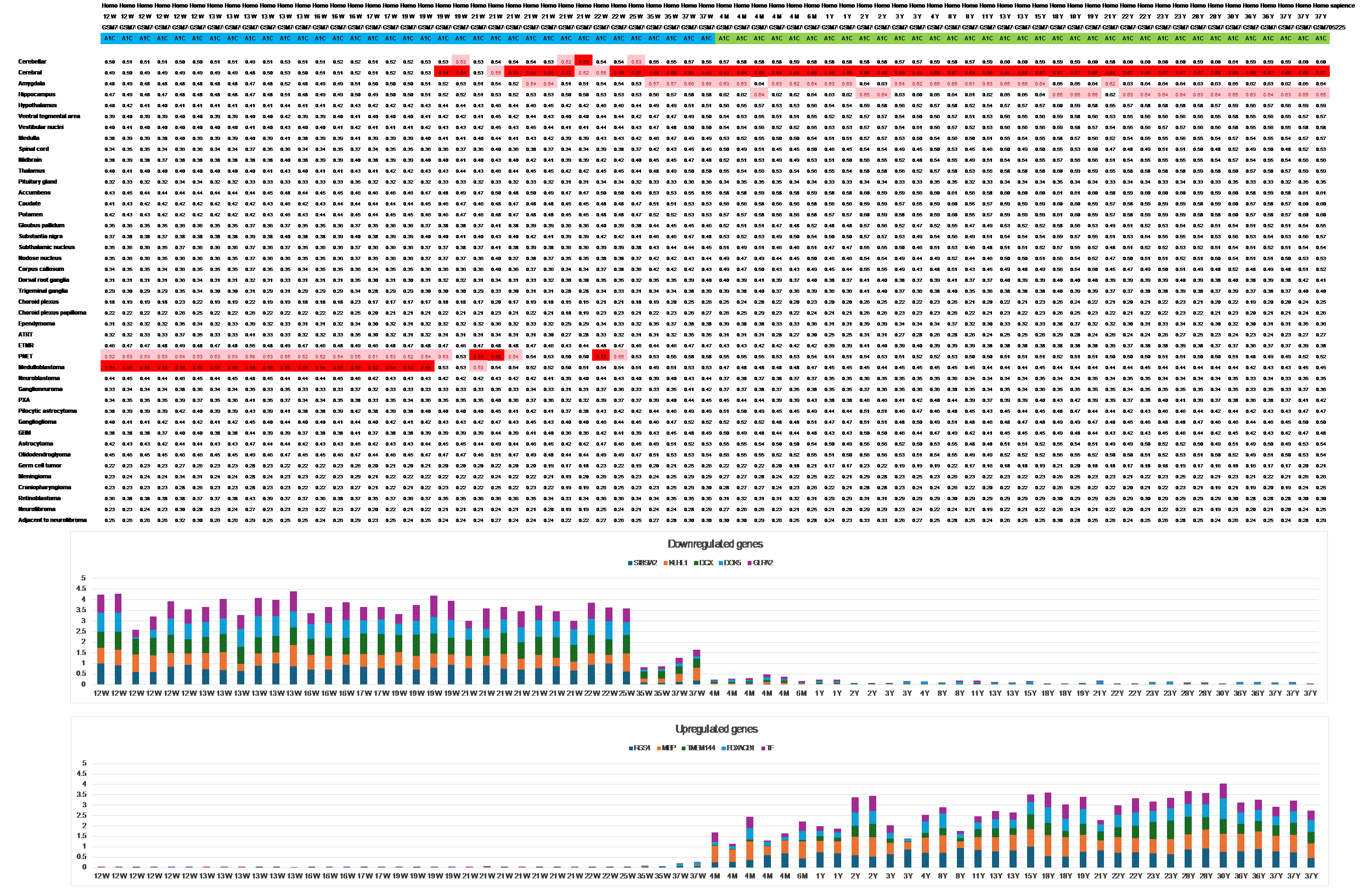
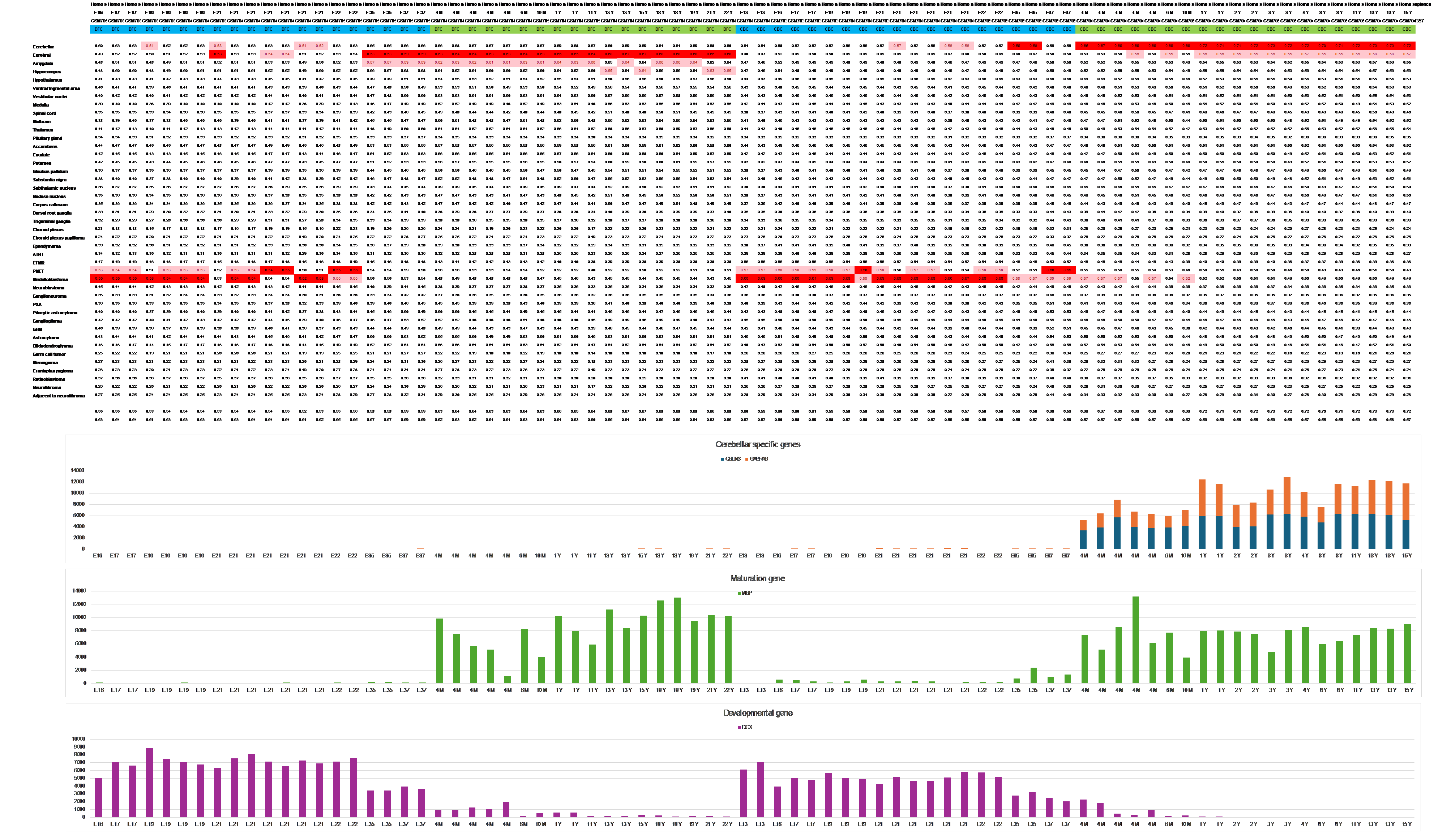
Observations from the “Spatio-temporal transcriptome of the human brain” series (GSE25219), as depicted in Figure 1, along with additional large human series available in Atlas II, reveal intriguing patterns.
Embryonic Expression Signature
Until mid-gestation, all human brain transcriptomes consistently exhibit an embryonic expression signature irrespective of the brain location.
This signature aligns with medulloblastoma or primitive neuroectodermal tumors (PNET).
Transition to Adult Expression Pattern:
Starting from gestational week 19, there is a shift from the primitive signature to the adult expression pattern of the same tissue.
The transition from the primitive to the mature expression signature is associated with the downregulation and upregulation of numerous genes.
Many of the downregulated genes are related to development, while the upregulated genes include those associated with myelination.
Figure 1 Until mid-gestation, the expression pattern closely resembles a primitive state, aligning with the transcriptomes of embryonic tumors. Starting from gestational week 19, there is a shift in individual specimens from the primitive to adult expression patterns, mirroring the same expression signature seen across different ages. Notably, the transition from the primitive to the mature expression signature appears to precede the downregulation and upregulation of the developmental and the maturation genes in this series (Primary auditory cortex, Dataset GSE25219)
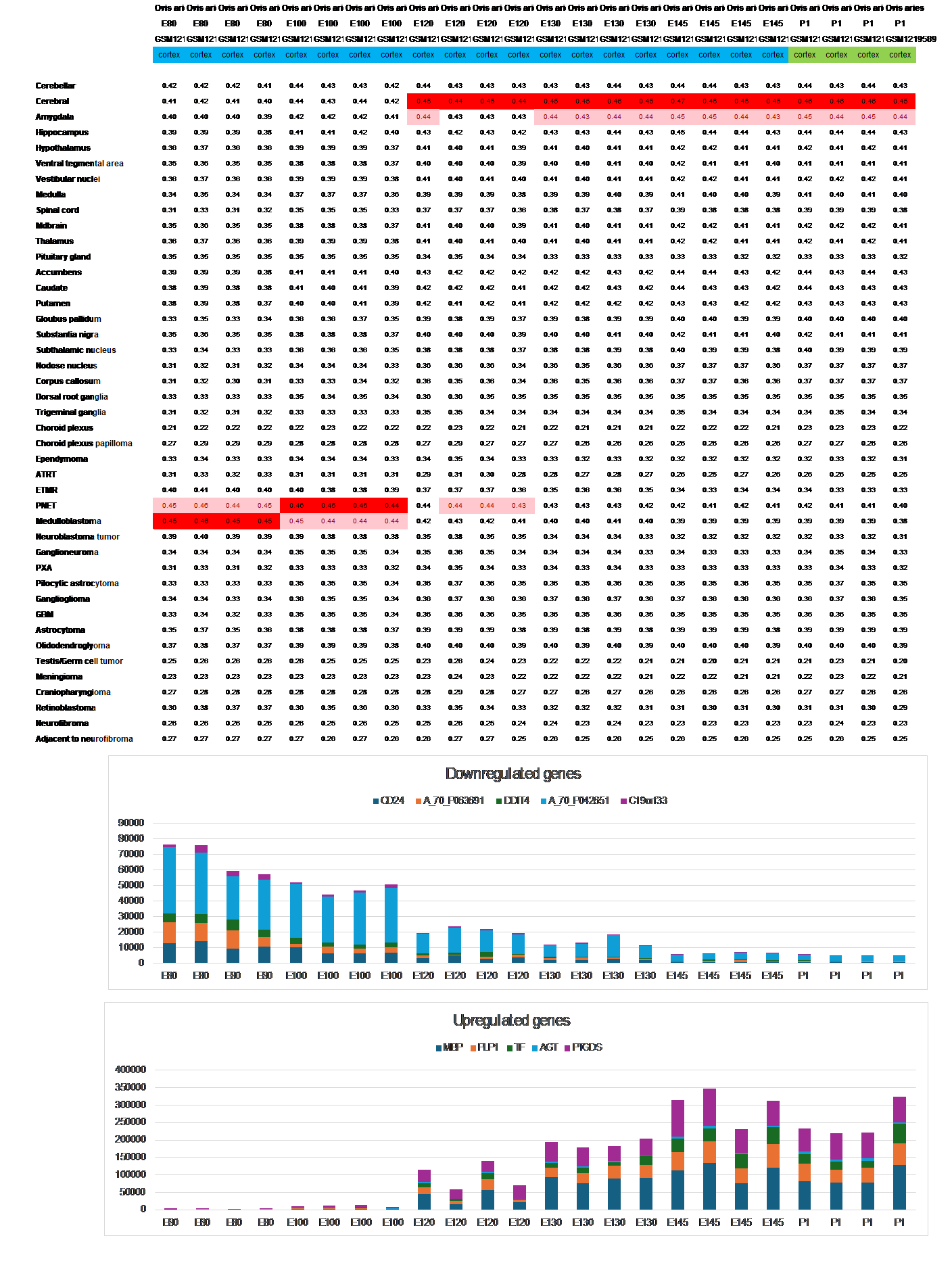
Similar to humans, the expression pattern of the sheep fetal brain begins aligning with the adult reference expression signature already from mid-pregnancy as observed in Figure 2 (Atlas III, dataset GSE50460).
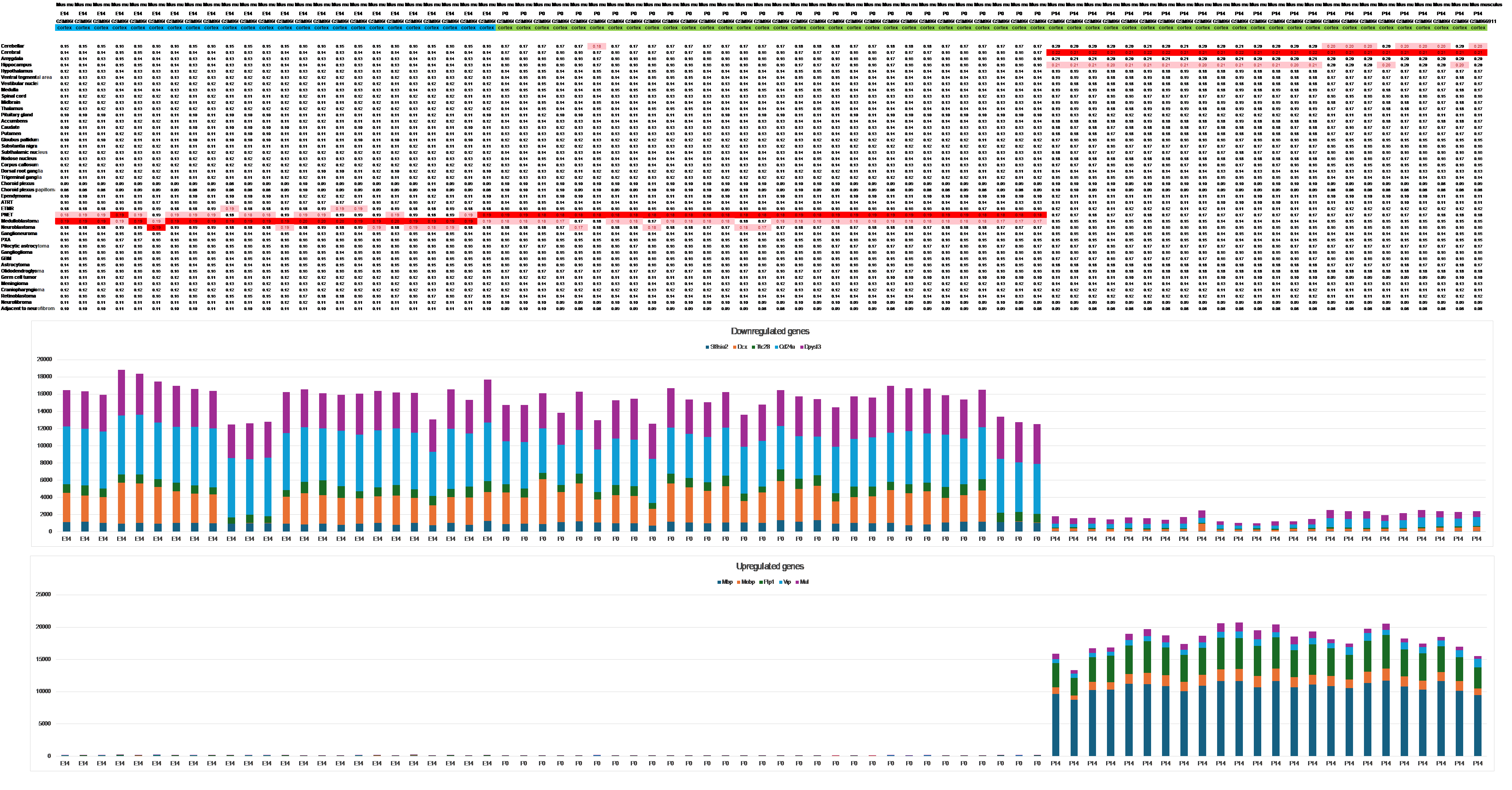
In contrast to humans and sheep, the expression pattern of mouse and rat brain transcriptomes consistently aligns with the reference expression signature of the aforementioned embryonic tumors throughout the entire pregnancy period and even during the first postpartum period (see Figure 3).
The transition from an ‘embryonic’ to an adult expression pattern during the early neonatal period occurs rapidly, spanning between postnatal days 9 (P9) and 11 (P11) in the mouse brain, as observed from dataset GSE62020. Similarly, in rats, this switch occurs between P7 and P14, as evidenced from GSE13793. This shift aligns with a postnatal differentiation wave in the already well-structured brains of these animals.
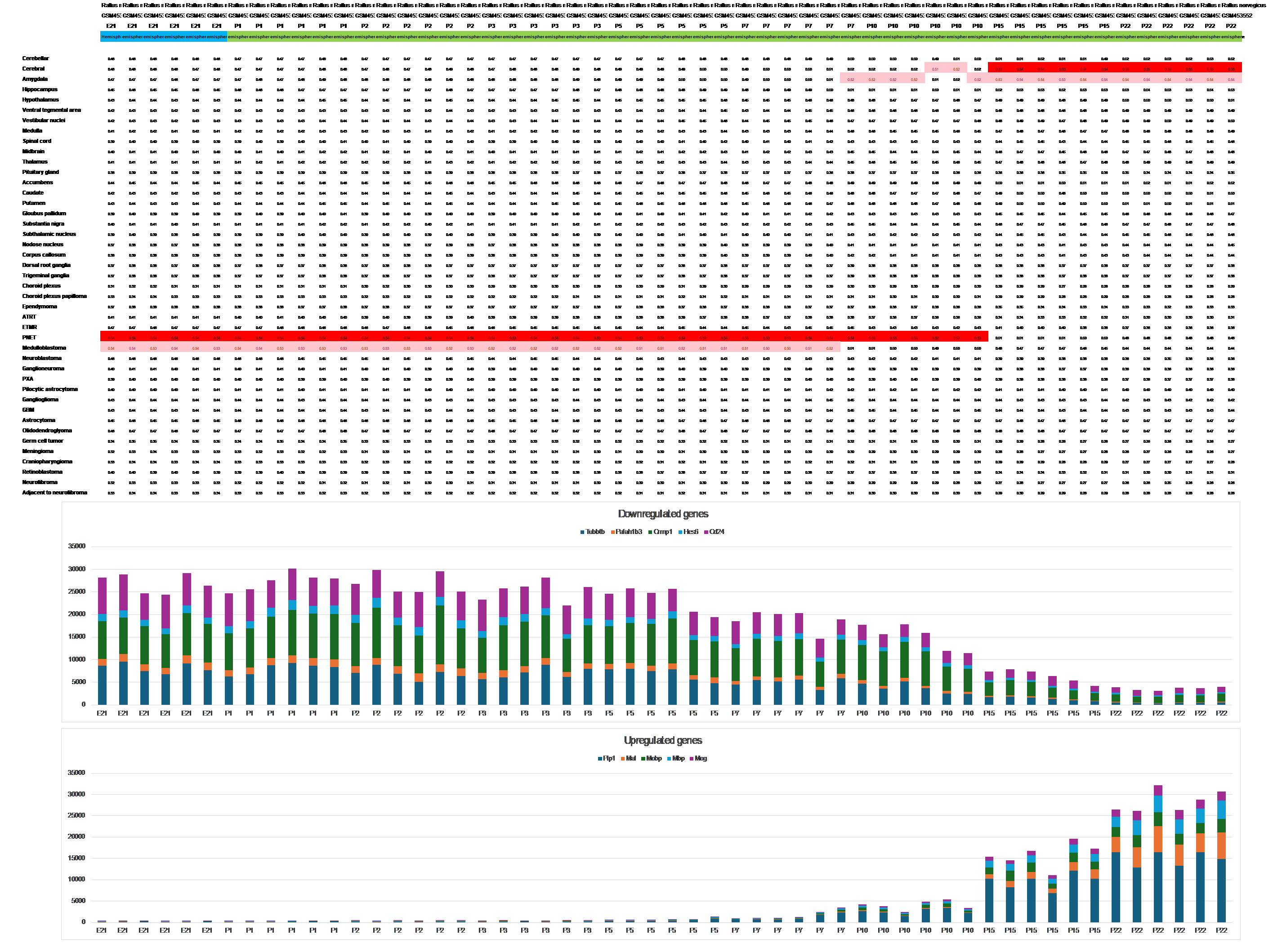
The entire maturation period of the rat brain, involving downregulation and upregulation of developmental and maturation genes, is accomplished within 3 weeks, with the first changes noticeable on day 5 postpartum (P5), as seen in Figure 4.
Figure 2 The switch from a primitive to a mature expression signature during mid-pregnancy in sheep is accompanied by the prompt upregulation of dozens of genes, including those associated with myelination, as well as downregulation of many developmental genes.
Figure 3 The switch from a primitive to a mature expression signature appears in mouse and rat brains only during the neonatal period, possibly due to the short pregnancy duration (cerebral cortex, GSE35366).
Figure 4 The first gene expression changes in the maturation of rat brain hemispheres can be observed from day 5 postpartum (P5) and are fully accomplished by P22, as observed from dataset GSE18133.
Brain Maturation and Tissue-Specific Genes
In addition to general maturation, which is reflected by the upregulation of maturation genes—such as myelin-related genes—across all brain regions during the transition from a primitive to a mature expression signature, there is also an intriguing upregulation of tissue-specific genes.
Notably, before the switch in expression, ‘cerebellar defining genes’ like CBLN3, GABRA6, and CRTAM, which are prominently overexpressed in the cerebellum compared to other brain regions, appear silent. This observation seems to reflect not only the generalized brain’s immaturity until late developmental stages but also implies its undifferentiated state in relation to tissue specificity.
As observed in Figure 5 and Figure 6, along with the switch in expression to an adult signature and the upregulation of maturation genes across all brain regions, some cerebellar-specific genes begin their overexpression during this late developmental stage in mice (Figure 5, dataset GSE47516) and in humans (Figure 6, dataset GSE13793). This dual pattern highlights both general maturation and tissue-specific terminal differentiation of the primitive cerebellar cells, leading to their final fate.
Figure 5 Upregulation of Cerebellar-Specific Genes during the Switch from a Primitive to an Adult Expression Signature in Cerebellar Mouse Transcriptomes (Dataset GSE47516)
This figure illustrates the dynamic expression changes of cerebellar-specific genes—CBLN3, GABRA6, MAL, and Gabra1—as the cerebellum transitions from a primitive to an adult expression profile. These genes play crucial roles in shaping the mature cerebellar structure.
Figure 6 In addition to the generalized upregulation of many maturation genes (such as Myelin basic protein, MBP) during the switch in expression to an adult signature, some of the prominent genes that distinguish cerebellar from cerebral tissues (specifically CBLN3 and GABRA6) become active. These genes are activated in the cerebellar cortex (CBC) but not in cerebral specimens from the dorsolateral prefrontal cortex (DFC), highlighting differential gene activation during this late time period.
Brain Maturation and Retinal Development
The persistence of the primitive expression pattern in mice and rat brains until their first neonatal stage corresponds to the delayed peak brain growth spurt and functional development compared to human newborns (6-9 weeks).
Similarly, the maturation of the mouse retina (which is also part of the central nervous system) appears to be delayed until the postpartum period, as observed in Atlas II (GSE24512, GSE74181, GSE33088).
Discussion
Unifying Transcriptomes Across Species and Platforms
This work, born from the collaborative efforts of scientists worldwide spanning over two decades, underscores the remarkable feasibility of analyzing transcriptomes across diverse series and species together. Importantly, it demonstrates that a single reference panel can effectively accommodate different platforms. Moreover, it highlights the advantage of comparing characteristic patterns of numerous genes, rather than focusing solely on a few highly expressed genes. The expression intensity differences across thousands of genes compensate for the small variations in expression intensity among most genes from different brain categories.
A pivotal message from this study lies in the continuous need for refining and enhancing the reference panel, potentially by incorporating additional series and molecular modalities, such as microRNA and methylome data.
In practical applications, leveraging the expression signature panel to analyze new test brain specimens can enhance diagnostic precision, particularly in cases where histopathology alone yields inconclusive results or to improve the uniformity of specimens for research or clinical trials. Moreover, the reference panel can serve as a valuable tool for quality control, ensuring that a given specimen genuinely aligns with other members within the same category. For instance:
Using the reference samples for brain tissues
As mentioned before, the reference panel showed variabilities in its matching with structures like amygdala and hippocampus across different series, where in some of them there is a great match (GSE62346 and GSE240873) while in other there is overlap among each other or with the cerebral reference signature. This raises doubt about whether the specimens included the same anatomical structures or were perhaps contaminated with cerebral tissue. In this respect, the similarity in expression signature may improve comparisons of brain tissues from different sources.
Using the reference panel for cell lines
While many cell lines in this meta-analysis exhibit strong correlations with their corresponding reference gene expression signature (notable examples include GBM-derived neurosphere cultures, glioblastoma stem-like cell lines (GSE23806), the neuroblastoma cell line IMR32 (GSE16254), the retinoblastoma cell line RB1 (GSE29683), the human brain neuronal cell line SH-SY5Y (GSE2732), the human GBM-derived stem-like cell line NCH421k (GSE134470), and the pediatric medulloblastoma cell line MB002 (GSE51020)), skepticism arises regarding certain cell lines and their fidelity in representing the original neoplastic clone.
For instance, the neuroblastoma cell line SKNAS-MYCN, derived from a 6-year-old White female patient with neuroblastoma (GSE16254), and the glioblastoma cell line GBM_Hs683 (GSE9171) both align best with the reference gene expression profile (GEP) of germ cell tumors.
Additionally, the neuroblastoma cell lines SK-NKAN, SK-N-SH, SMS-SAN, SH-SY5Y, and NB-16 all align best with atypical teratoid/rhabdoid tumor (ATRT) in GSE78061, not with the reference panel of neuroblastoma. All the above instances are from Atlas I.
Brain Development and Expression Signatures
The persistence of the primitive expression signature until mid-pregnancy in humans and sheep, and until the early postnatal life in mice and rats, along with the subsequent switch to the mature expression signature, suggests an intriguing parallel to the primary cartilage model of limb element development [10]. Until mid-pregnancy in humans and sheep (and possibly other primates and large mammals), and until the first neonatal period in small rodents, the fetal brain appears as a three-dimensional model composed of primitive neuroectoderm.
Only after achieving its final structural organization does it embark on maturation toward specific cell fates, accompanied by the loss of migration, proliferation, and other developmental capabilities.
In practice, familiarity with the timing of the switch in expression from the primitive to the adult signature can direct investigations to this critical period and facilitate the study of brain development. Notably, many animal investigations conducted during the early neonatal periods were based on brain specimens that actually expressed an embryonic expression signature. Conversely, fetal development investigations may be equally effective if conducted in mice and rats during the first neonatal week.
Finally, from a philosophical or ethical standpoint, the transition from a primitive to mature pattern of the human brain expression signature beyond gestational week 19 may signify a point where a primitive brain model acquires an indistinguishable pattern from the adult brain.
Future Directions
Ongoing efforts are directed toward expanding the reference panel to additional organs, the next project is bone marrow including all available transcriptome datasets. The integration of expression signature for each tissue type with genomic and complementary molecular data may be useful in the future for providing a precise diagnosis by itself, giving more objective measurements to histological diagnosis.
Supplementary Methods
In my preliminary reference panel, I tested all 54,614 probes from the GPL570 microarray. However, this approach resulted in excessive overlap among different brain tissue categories. Upon closer examination of probelevel intensity, I observed that while many probes showed consistent expression intensity, others did not. Furthermore, the expression intensity of many probes is generally very high, and their intensities span a wide range. Such probes consistently exhibit significant differences among different specimens, even if those specimens share the same histology. Additionally, these high-intensity probes tend to overshadow the contributions of other probes that exhibit smaller but consistent differences across various histological brain categories.
To address this issue, I established the 800-5 criteria for selecting probes to construct the reference GEPs. Specifically, I chose probes where the difference in signal intensity among different representative GEPs exceeded 800, while also ensuring that the difference was at least 5 times greater. For example:
If the average expression of the FOS probe in a representative cerebellar series was 1200, compared to 50 in a representative cerebral series, the FOS probe was included in the reference list.
Conversely, if the respective intensities were 3000 vs. 1500, the FOS probe was removed, even though the absolute difference was greater in the latter case.
Ultimately, I conducted pairwise comparisons among the 43 representative GEPs, resulting in a total of 1806 comparisons. This rigorous process yielded 4621 transcripts whose expression met the stringent 800–5 criterion.
Conflict of Interest
The author declares no conflict of interest.
Supplementary data are available for download at
https://brain-cell-of-origin-atlas.website- Visvader JE (2011) Cells of origin in cancer. Nature. 469: 7330.
- Alizadeh AA, Eisen MB, Davis RE, Ma C, Lossos IS, Rosenwald A, Boldrick JC, et al. (2000) Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature. 403: 503-11.
- Zhan F, Huang Y, Colla S, Stewart JP, Hanamura I, Gupta S, et al. (2006) The molecular classification of multiple myeloma. Blood. 108: 2020-8.
- Kim HJ, Park JW, Jeong HL (2021) Genetic Architectures and Cell of Origin in Glioblastoma. Front Oncol.
- Lee JH, Lee JH (2018) The origin-of-cell harboring cancer-driving mutations in human glioblastoma. MB Rep. 51: 481-3.
- Alcantara Llaguno SR, Parada LF (2016) Cell of origin of glioma: biological and clinical implications. Br J Cancer. 115: 1445-50.
- Zhang W, Zhang Y, Zheng Y, Zheng M, Sun N, Yang X, et al. (2019) Progress in Research on Brain Development and Function of Mice During Weaning. Curr Protein Pept Sci. 20: 705-12.
- Pressler R, Stéphane Auvin S (2013) Comparison of Brain Maturation among Species: An Example in Translational Research Suggesting the Possible Use of Bumetanide in Newborn. Front Neurol. 4: 36.
- Zeiss CJ (2021) Comparative Milestones in Rodent and Human Postnatal Central Nervous System Development. Toxicol Pathol. 49: 1368-73.
- DeLise AM, Fischer L, Tuan RS (2000) Cellular interactions and signaling in cartilage development. Osteoarthritis Cartilage. 8: 309-34.


Tables at a glance
Figures at a glance